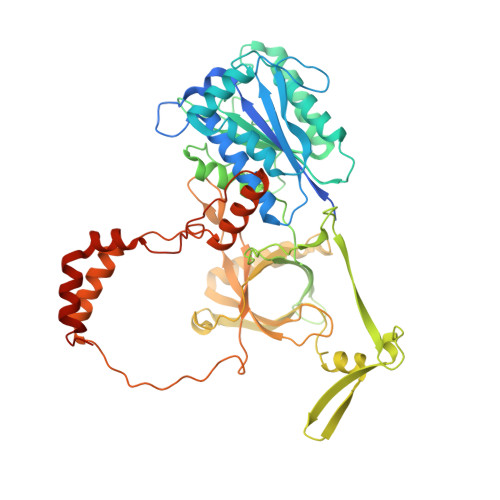
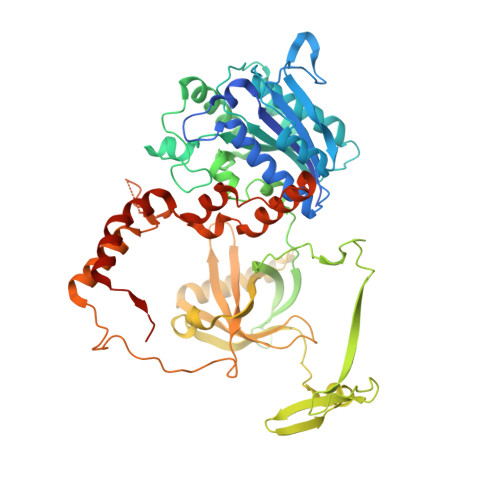
PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Seif-El-Dahan, M., Kefala-Stavridi, A., Frit, P., Hardwick, S.W., Chirgadze, D.Y., Maia De Oliviera, T., Britton, S., Barboule, N., Bossaert, M., Pandurangan, A.P., Meek, K., Blundell, T.L., Ropars, V., Calsou, P., Charbonnier, J.B., Chaplin, A.K.(2023) Sci Adv 9: eadg2834-eadg2834
- PubMed: 37256950
- DOI: https://doi.org/10.1126/sciadv.adg2834
- Primary Citation of Related Structures:
7ZWA, 7ZYG, 8ASC, 8BH3, 8BHV, 8BHY - PubMed Abstract:
Nonhomologous end joining is a critical mechanism that repairs DNA double-strand breaks in human cells. In this work, we address the structural and functional role of the accessory protein PAXX [paralog of x-ray repair cross-complementing protein 4 (XRCC4) and XRCC4-like factor (XLF)] in this mechanism. Here, we report high-resolution cryo-electron microscopy (cryo-EM) and x-ray crystallography structures of the PAXX C-terminal Ku-binding motif bound to Ku70/80 and cryo-EM structures of PAXX bound to two alternate DNA-dependent protein kinase (DNA-PK) end-bridging dimers, mediated by either Ku80 or XLF. We identify residues critical for the Ku70/PAXX interaction in vitro and in cells. We demonstrate that PAXX and XLF can bind simultaneously to the Ku heterodimer and act as structural bridges in alternate forms of DNA-PK dimers. Last, we show that engagement of both proteins provides a complementary advantage for DNA end synapsis and end joining in cells.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), Institute Joliot, CEA, CNRS, Université Paris-Saclay, 91198 Gif-sur-Yvette cedex, France.