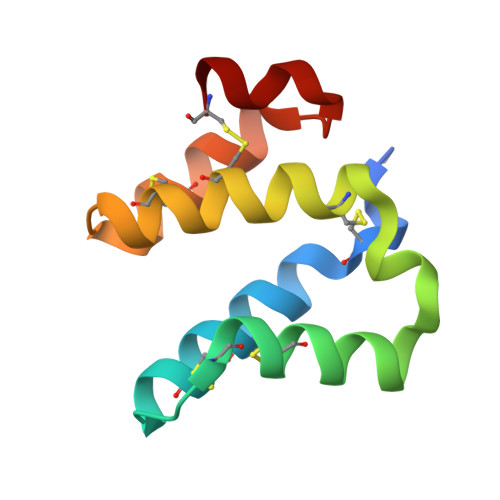
High-resolution crystal structure of the Mu8.1 conotoxin from Conus mucronatus.
Muller, E., Hackney, C.M., Ellgaard, L., Morth, J.P.(2023) Acta Crystallogr F Struct Biol Commun 79: 240-246
- PubMed: 37642664
- DOI: https://doi.org/10.1107/S2053230X23007070
- Primary Citation of Related Structures:
8AMY - PubMed Abstract:
Marine cone snails produce a wealth of peptide toxins (conotoxins) that bind their molecular targets with high selectivity and potency. Therefore, conotoxins constitute valuable biomolecular tools with a variety of biomedical purposes. The Mu8.1 conotoxin from Conus mucronatus is the founding member of the newly identified saposin-like conotoxin class of conotoxins and has been shown to target Cav2.3, a voltage-gated calcium channel. Two crystal structures have recently been determined of Mu8.1 at 2.3 and 2.1 Å resolution. Here, a high-resolution crystal structure of Mu8.1 was determined at 1.67 Å resolution in the high-symmetry space group I4 1 22. The asymmetric unit contained one molecule, with a symmetry-related molecule generating a dimer equivalent to that observed in the two previously determined structures. The high resolution allows a detailed atomic analysis of a water-filled cavity buried at the dimer interface, revealing a tightly coordinated network of waters that shield a lysine residue (Lys55) with a predicted unusually low side-chain pK a value. These findings are discussed in terms of a potential functional role of Lys55 in target interaction.
Organizational Affiliation:
Department of Biotechnology and Biomedicine, Technical University of Denmark, 2800 Kongens Lyngby, Denmark.