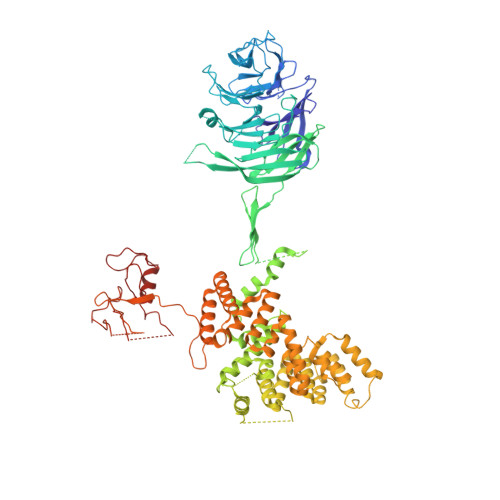
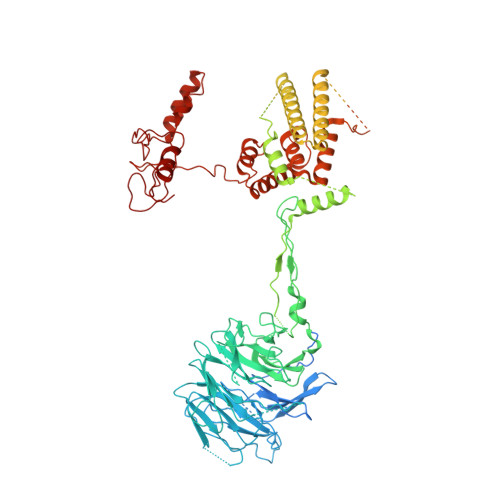
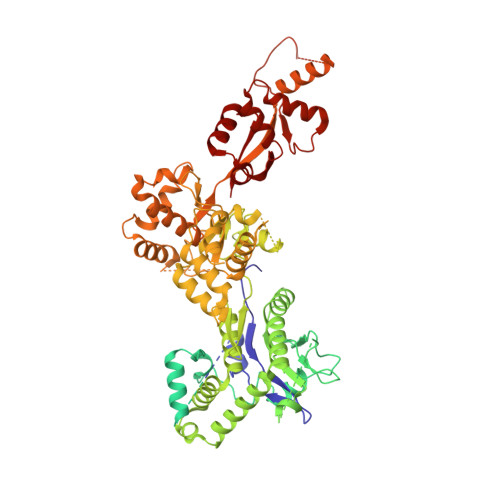
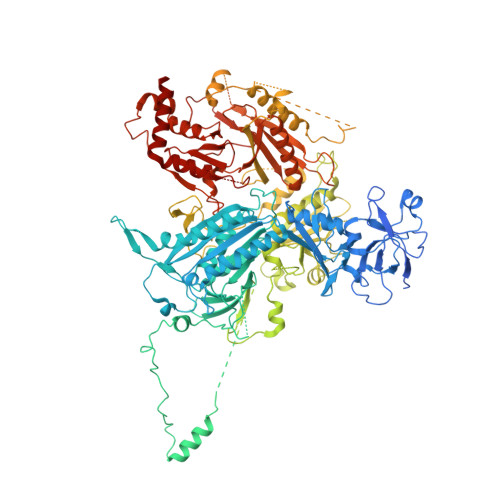
Cryo-EM structure of the SEA complex.
Tafur, L., Hinterndorfer, K., Gabus, C., Lamanna, C., Bergmann, A., Sadian, Y., Hamdi, F., Kyrilis, F.L., Kastritis, P.L., Loewith, R.(2022) Nature 611: 399-404
- PubMed: 36289347
- DOI: https://doi.org/10.1038/s41586-022-05370-0
- Primary Citation of Related Structures:
8ADL, 8AE6 - PubMed Abstract:
The SEA complex (SEAC) is a growth regulator that acts as a GTPase-activating protein (GAP) towards Gtr1, a Rag GTPase that relays nutrient status to the Target of Rapamycin Complex 1 (TORC1) in yeast 1 . Functionally, the SEAC has been divided into two subcomplexes: SEACIT, which has GAP activity and inhibits TORC1, and SEACAT, which regulates SEACIT 2 . This system is conserved in mammals: the GATOR complex, consisting of GATOR1 (SEACIT) and GATOR2 (SEACAT), transmits amino acid 3 and glucose 4 signals to mTORC1. Despite its importance, the structure of SEAC/GATOR, and thus molecular understanding of its function, is lacking. Here, we solve the cryo-EM structure of the native eight-subunit SEAC. The SEAC has a modular structure in which a COPII-like cage corresponding to SEACAT binds two flexible wings, which correspond to SEACIT. The wings are tethered to the core via Sea3, which forms part of both modules. The GAP mechanism of GATOR1 is conserved in SEACIT, and GAP activity is unaffected by SEACAT in vitro. In vivo, the wings are essential for recruitment of the SEAC to the vacuole, primarily via the EGO complex. Our results indicate that rather than being a direct inhibitor of SEACIT, SEACAT acts as a scaffold for the binding of TORC1 regulators.
- Department of Molecular and Cellular Biology, University of Geneva, Geneva, Switzerland.
Organizational Affiliation: