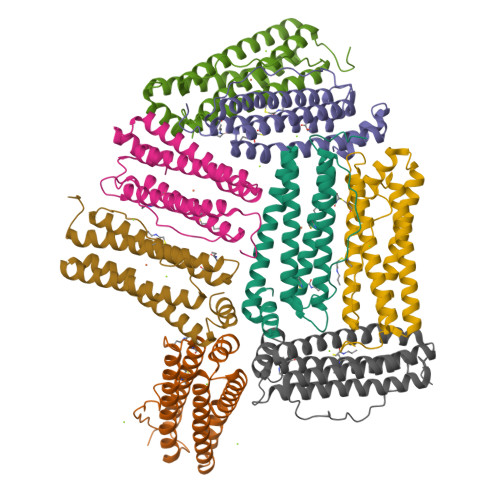
Assembly of chemically modified protein nanocages into 3D materials for the adsorption of uremic toxins.
Bohler, H., Orth-Alampour, S., Baaten, C., Riedner, M., Jankowski, J., Beck, T.(2022) J Mater Chem B 11: 55-60
- PubMed: 36504125
- DOI: https://doi.org/10.1039/d2tb02386e
- Primary Citation of Related Structures:
8AAV - PubMed Abstract:
Hemodialysis fails to remove protein-bound uremic toxins that are attributed with high cardiovascular risk. Application of adsorption materials is a viable strategy, but suitable biocompatible adsorbents are still not available. Here, we demonstrate that adsorbents based on the bottom-up assembly of the intrinsically biocompatible protein cage ferritin are applicable for toxin adsorption. Due to the size-exclusion effect of its pores, only small molecules such as uremic toxins can enter the protein cage. Protein redesign techniques that target selectively the inner surface were used to introduce anchor sites for chemical modification. Porous crystalline adsorbents were fabricated by bottom-up assembly of the protein cage. Linkage of up to 96 phenylic or aliphatic molecules per container was verified by ESI-MS. Materials based on unmodified ferritin cages can already adsorb the uremic toxins. The adsorption capacity could be increased by about 50% through functionalization with hydrophobic molecules reaching 458 μg g -1 for indoxyl sulfate. The biohybrid materials show no contamination with endotoxins and do not activate blood platelets. These findings demonstrate the great potential of protein-based adsorbents for the clearance of uremic toxins: modifications enhance toxin adsorption without diminishing the biocompatibility of the final protein-based material.
Organizational Affiliation:
Universität Hamburg, Department of Chemistry, Institute of Physical Chemistry, Grindelallee 117, Hamburg 20146, Germany. tobias.beck@chemie.uni-hamburg.de.