Sebetralstat (KVD900): A Potent and Selective Small Molecule Plasma Kallikrein Inhibitor Featuring a Novel P1 Group as a Potential Oral On-Demand Treatment for Hereditary Angioedema.
Davie, R.L., Edwards, H.J., Evans, D.M., Hodgson, S.T., Stocks, M.J., Smith, A.J., Rushbrooke, L.J., Pethen, S.J., Roe, M.B., Clark, D.E., McEwan, P.A., Hampton, S.L.(2022) J Med Chem 65: 13629-13644
- PubMed: 36251573
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00921
- Primary Citation of Related Structures:
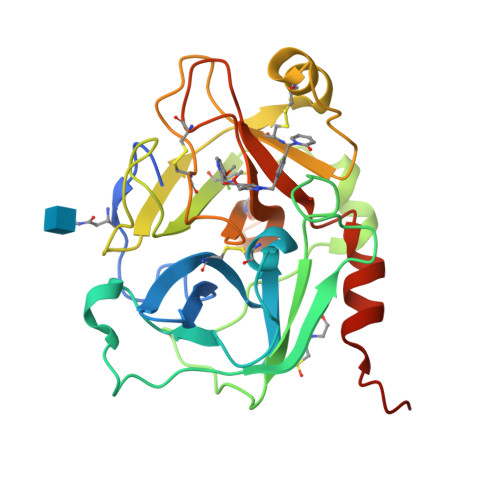
8A3Q - PubMed Abstract:
Hereditary angioedema (HAE) is a rare genetic disorder in which patients experience sudden onset of swelling in various locations of the body. HAE is associated with uncontrolled plasma kallikrein (PKa) enzyme activity and generation of the potent inflammatory mediator, bradykinin, resulting in episodic attacks of angioedema. Herein, we disclose the discovery and optimization of novel small molecule PKa inhibitors. Starting from molecules containing highly basic P1 groups, which typically bind to an aspartic acid residue (Asp189) in the serine protease S1 pocket, we identified novel P1 binding groups likely to have greater potential for oral-drug-like properties. The optimization of P4 and the central core together with the particularly favorable properties of 3-fluoro-4-methoxypyridine P1 led to the development of sebetralstat, a potent, selective, orally bioavailable PKa inhibitor in phase 3 for on-demand treatment of HAE attacks.
Organizational Affiliation:
KalVista Pharmaceuticals Limited, Porton Science Park, Salisbury, SP4 0BF, U.K.