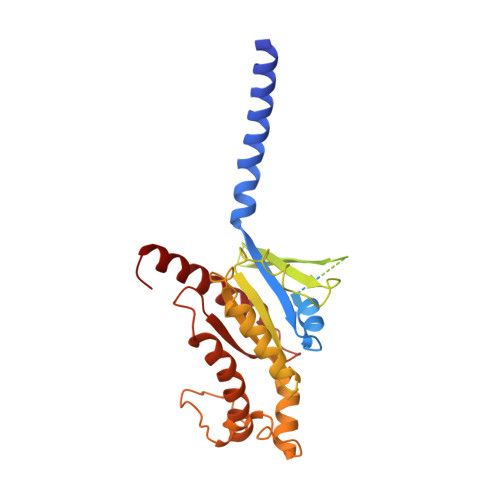
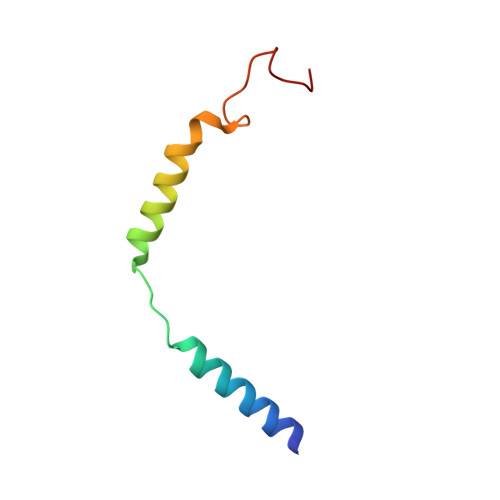
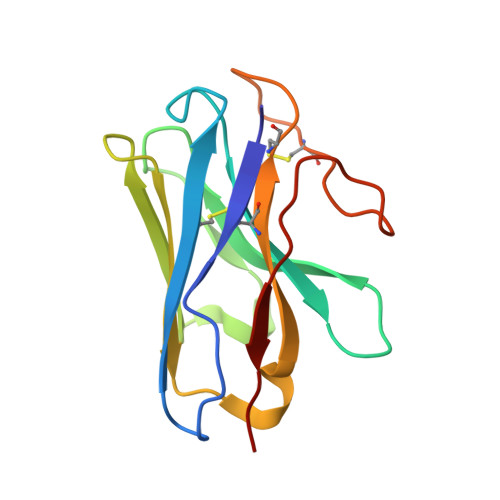
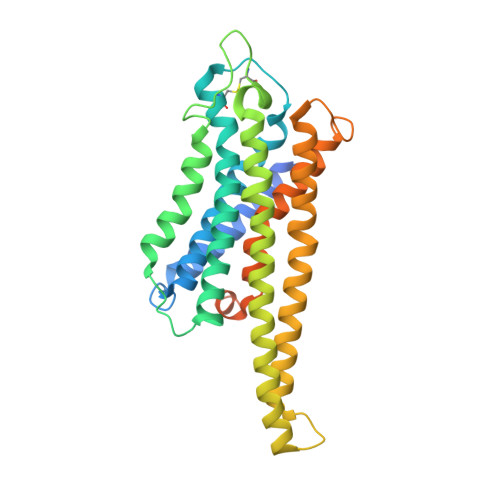
Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Ma, L., Yang, F., Wu, X., Mao, C., Guo, L., Miao, T., Zang, S.K., Jiang, X., Shen, D.D., Wei, T., Zhou, H., Wei, Q., Li, S., Shu, Q., Feng, S., Jiang, C., Chu, B., Du, L., Sun, J.P., Yu, X., Zhang, Y., Zhang, P.(2022) Proc Natl Acad Sci U S A 119: e2117054119-e2117054119
- PubMed: 35858343
- DOI: https://doi.org/10.1073/pnas.2117054119
- Primary Citation of Related Structures:
7XTQ - PubMed Abstract:
The G protein-coupled bile acid receptor (GPBAR) is the membrane receptor for bile acids and a driving force of the liver-bile acid-microbiota-organ axis to regulate metabolism and other pathophysiological processes. Although GPBAR is an important therapeutic target for a spectrum of metabolic and neurodegenerative diseases, its activation has also been found to be linked to carcinogenesis, leading to potential side effects. Here, via functional screening, we found that two specific GPBAR agonists, R399 and INT-777, demonstrated strikingly different regulatory effects on the growth and apoptosis of non-small cell lung cancer (NSCLC) cells both in vitro and in vivo. Further mechanistic investigation showed that R399-induced GPBAR activation displayed an obvious bias for β-arrestin 1 signaling, thus promoting YAP signaling activation to stimulate cell proliferation. Conversely, INT-777 preferentially activated GPBAR-Gs signaling, thus inactivating YAP to inhibit cell proliferation and induce apoptosis. Phosphorylation of GPBAR by GRK2 at S310/S321/S323/S324 sites contributed to R399-induced GPBAR-β-arrestin 1 association. The cryoelectron microscopy (cryo-EM) structure of the R399-bound GPBAR-Gs complex enabled us to identify key interaction residues and pivotal conformational changes in GPBAR responsible for the arrestin signaling bias and cancer cell proliferation. In summary, we demonstrate that different agonists can regulate distinct functions of cell growth and apoptosis through biased GPBAR signaling and control of YAP activity in a NSCLC cell model. The delineated mechanism and structural basis may facilitate the rational design of GPBAR-targeting drugs with both metabolic and anticancer benefits.
- Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan 250012, China.
Organizational Affiliation: