Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Alhashimi, R.T., Ghatge, M.S., Donkor, A.K., Deshpande, T.M., Anabaraonye, N., Alramadhani, D., Danso-Danquah, R., Huang, B., Zhang, Y., Musayev, F.N., Abdulmalik, O., Safo, M.K.(2022) Biomolecules 12
- PubMed: 35625623
- DOI: https://doi.org/10.3390/biom12050696
- Primary Citation of Related Structures:
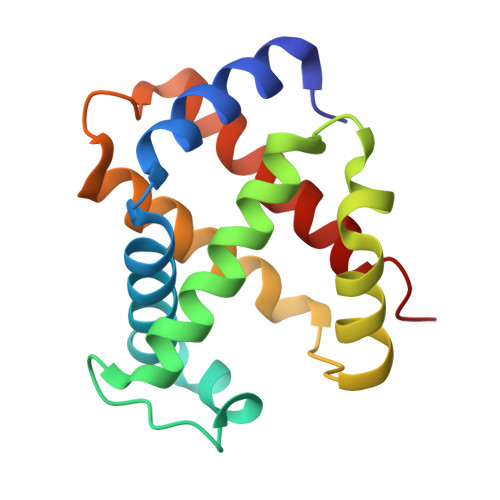
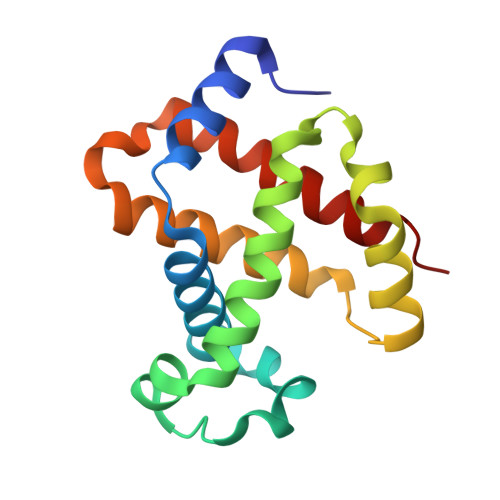
7UD7, 7UD8 - PubMed Abstract:
5-hydroxyfurfural (5HMF), an allosteric effector of hemoglobin (Hb) with an ability to increase Hb affinity for oxygen has been studied extensively for its antisickling effect in vitro and in vivo, and in humans for the treatment of sickle cell disease (SCD). One of the downstream pathophysiologies of SCD is nitric oxide (NO) deficiency, therefore increasing NO (bio)availability is known to mitigate the severity of SCD symptoms. We report the synthesis of an NO-releasing prodrug of 5HMF (5HMF-NO), which in vivo, is expected to be bio-transformed into 5HMF and NO, with concomitant therapeutic activities. In vitro studies showed that when incubated with whole blood, 5HMF-NO releases NO, as anticipated. When incubated with sickle blood, 5HMF-NO formed Schiff base adduct with Hb, increased Hb affinity for oxygen, and prevented hypoxia-induced erythrocyte sickling, which at 1 mM concentration were 16%, 10% and 27%, respectively, compared to 21%, 18% and 21% for 5HMF. Crystal structures of 5HMF-NO with Hb showed 5HMF-NO bound to unliganded (deoxygenated) Hb, while the hydrolyzed product, 5HMF bound to liganded (carbonmonoxy-ligated) Hb. Our findings from this proof-of-concept study suggest that the incorporation of NO donor group to 5HMF and analogous molecules could be a novel beneficial strategy to treat SCD and warrants further detailed in vivo studies.
- Department of Medicinal Chemistry and The Institute for Structural Biology, Drug Discovery and Development, School of Pharmacy, Virginia Commonwealth University, Richmond, VA 23298, USA.
Organizational Affiliation: