Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Woloszyn, K., Vecchioni, S., Ohayon, Y.P., Lu, B., Ma, Y., Huang, Q., Zhu, E., Chernovolenko, D., Markus, T., Jonoska, N., Mao, C., Seeman, N.C., Sha, R.(2022) Adv Mater 34: e2206876-e2206876
- PubMed: 36100349
- DOI: https://doi.org/10.1002/adma.202206876
- Primary Citation of Related Structures:
7U3O, 7U3P, 7U3Q, 7U3R, 7U3S, 7U3T, 7U3U, 7U3V, 7U3W, 7U3X, 7U3Y, 7U3Z, 7U40, 7U41, 7U42, 7U43, 7U44, 7U45 - PubMed Abstract:
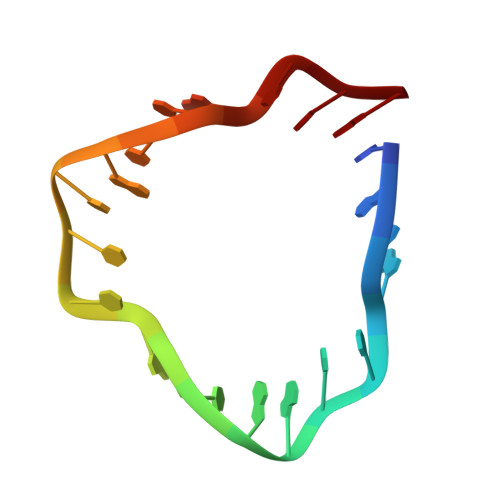
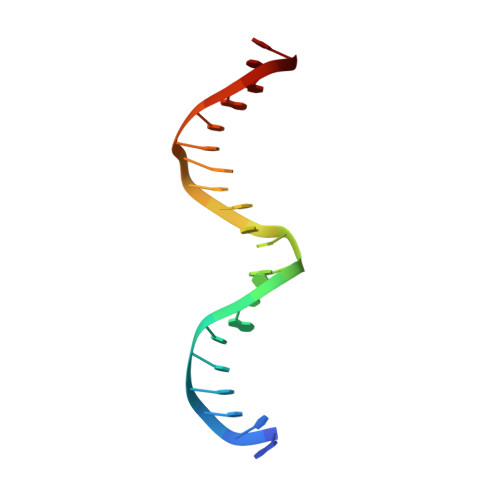
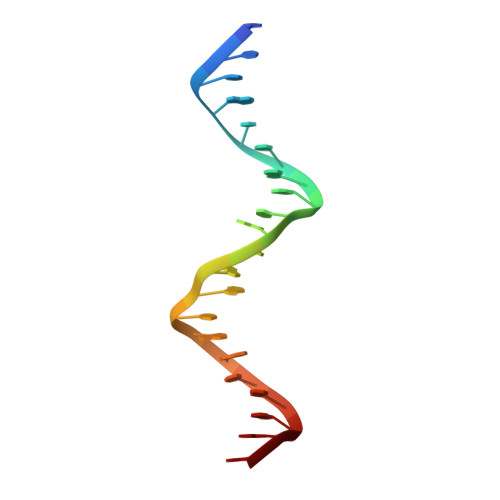
The DNA tensegrity triangle is known to reliably self-assemble into a 3D rhombohedral crystalline lattice via sticky-end cohesion. Here, the library of accessible motifs is expanded through covalent extensions of intertriangle regions and sticky-end-coordinated linkages of adjacent triangles with double helical segments using both geometrically symmetric and asymmetric configurations. The molecular structures of 18 self-assembled architectures at resolutions of 3.32-9.32 Å are reported; the observed cell dimensions, cavity sizes, and cross-sectional areas agree with theoretical expectations. These data demonstrate that fine control over triclinic and rhombohedral crystal parameters and the customizability of more complex 3D DNA lattices are attainable via rational design. It is anticipated that augmented DNA architectures may be fine-tuned for the self-assembly of designer nanocages, guest-host complexes, and proscriptive 3D nanomaterials, as originally envisioned. Finally, designer asymmetric crystalline building blocks can be seen as a first step toward controlling and encoding information in three dimensions.
Organizational Affiliation:
Department of Chemistry, New York University, New York, NY, 10003, USA.