Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Gibbs, E., Klemm, E., Seiferth, D., Kumar, A., Ilca, S.L., Biggin, P.C., Chakrapani, S.(2023) Nat Commun 14: 1363
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2023) Nat Commun 14: 1363
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycine receptor subunit alphaZ1 | A, B [auth D], C, D [auth B] | 458 | Danio rerio | Mutation(s): 0 Gene Names: glra1 Membrane Entity: Yes | 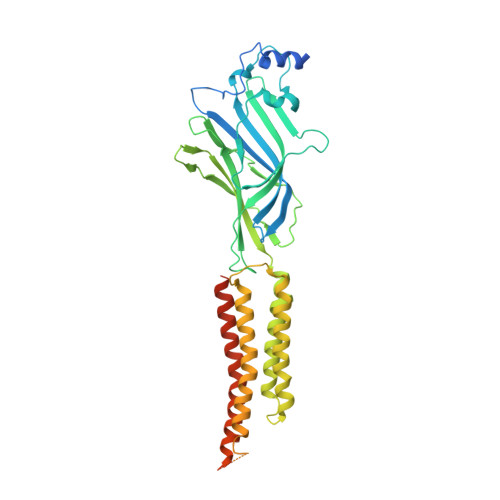 |
UniProt | |||||
Find proteins for O93430 (Danio rerio) Explore O93430 Go to UniProtKB: O93430 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O93430 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycine receptor beta subunit 2 | 591 | Danio rerio | Mutation(s): 0 Gene Names: glrbb, glrb2 Membrane Entity: Yes | 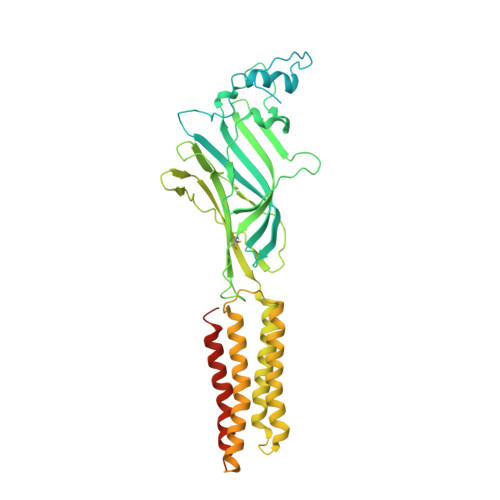 | |
UniProt | |||||
Find proteins for Q6DC22 (Danio rerio) Explore Q6DC22 Go to UniProtKB: Q6DC22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6DC22 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PIO Query on PIO | AA [auth B] G [auth A] HA [auth E] IA [auth E] J [auth A] | [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate C25 H49 O19 P3 XLNCEHRXXWQMPK-MJUMVPIBSA-N |  | ||
| PX4 Query on PX4 | BA [auth B] CA [auth B] DA [auth E] EA [auth E] H [auth A] | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE C36 H73 N O8 P CITHEXJVPOWHKC-UUWRZZSWSA-O |  | ||
| SY9 (Subject of Investigation/LOI) Query on SY9 | I [auth A], L [auth D], T [auth C], W [auth B], X [auth B] | STRYCHNINE C21 H22 N2 O2 QMGVPVSNSZLJIA-FVWCLLPLSA-N |  | ||
| NAG Query on NAG | F [auth A] FA [auth E] GA [auth E] K [auth D] S [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC | 3.3.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 5R35GM134896 |