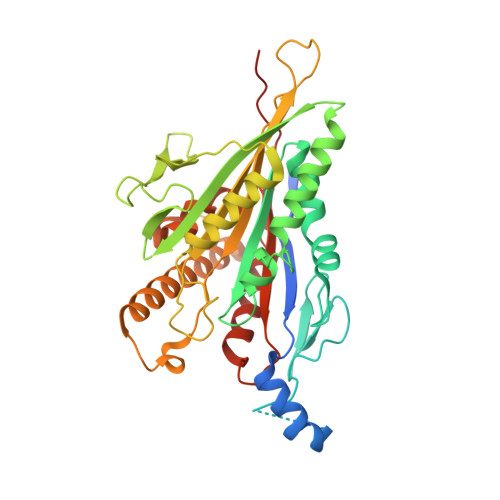
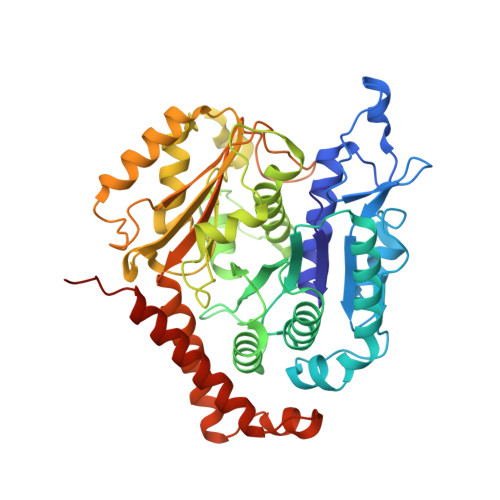
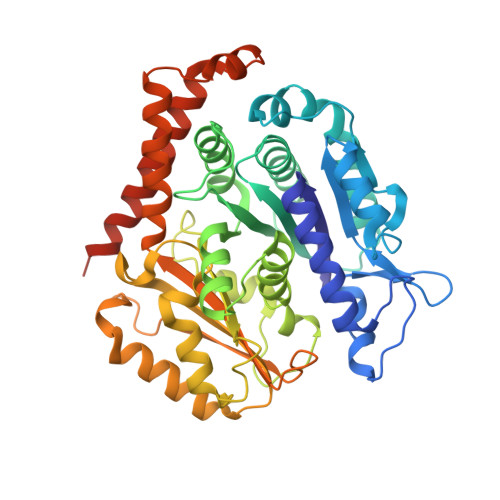
Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape.
Hunter, B., Benoit, M.P.M.H., Asenjo, A.B., Doubleday, C., Trofimova, D., Frazer, C., Shoukat, I., Sosa, H., Allingham, J.S.(2022) Nat Commun 13: 4198-4198
- PubMed: 35859148
- DOI: https://doi.org/10.1038/s41467-022-31794-3
- Primary Citation of Related Structures:
7LFF, 7TQX, 7TQY, 7TQZ, 7TR0, 7TR1, 7TR2, 7TR3 - PubMed Abstract:
Kinesin-8s are dual-activity motor proteins that can move processively on microtubules and depolymerize microtubule plus-ends, but their mechanism of combining these distinct activities remains unclear. We addressed this by obtaining cryo-EM structures (2.6-3.9 Å) of Candida albicans Kip3 in different catalytic states on the microtubule lattice and on a curved microtubule end mimic. We also determined a crystal structure of microtubule-unbound CaKip3-ADP (2.0 Å) and analyzed the biochemical activity of CaKip3 and kinesin-1 mutants. These data reveal that the microtubule depolymerization activity of kinesin-8 originates from conformational changes of its motor core that are amplified by dynamic contacts between its extended loop-2 and tubulin. On curved microtubule ends, loop-1 inserts into preceding motor domains, forming head-to-tail arrays of kinesin-8s that complement loop-2 contacts with curved tubulin and assist depolymerization. On straight tubulin protofilaments in the microtubule lattice, loop-2-tubulin contacts inhibit conformational changes in the motor core, but in the ADP-Pi state these contacts are relaxed, allowing neck-linker docking for motility. We propose that these tubulin shape-induced alternations between pro-microtubule-depolymerization and pro-motility kinesin states, regulated by loop-2, are the key to the dual activity of kinesin-8 motors.
- Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON, K7L 3N6, Canada.
Organizational Affiliation: