Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Santos, M.F.A., Sciortino, G., Correia, I., Fernandes, A.C.P., Santos-Silva, T., Pisanu, F., Garribba, E., Costa Pessoa, J.(2022) Chemistry 28: e202200105-e202200105
- PubMed: 35486702
- DOI: https://doi.org/10.1002/chem.202200105
- Primary Citation of Related Structures:
7Q0T, 7Q0U, 7Q0V, 7Q0W, 7Q0X - PubMed Abstract:
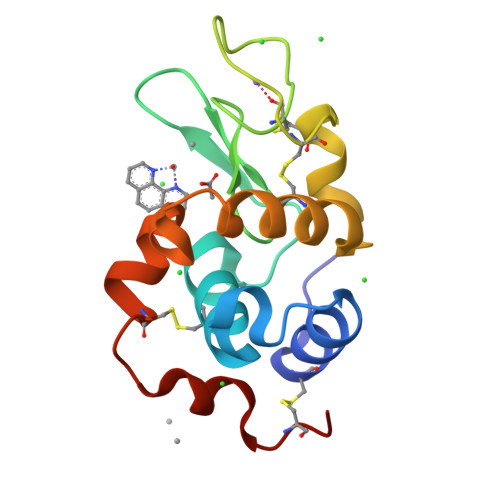
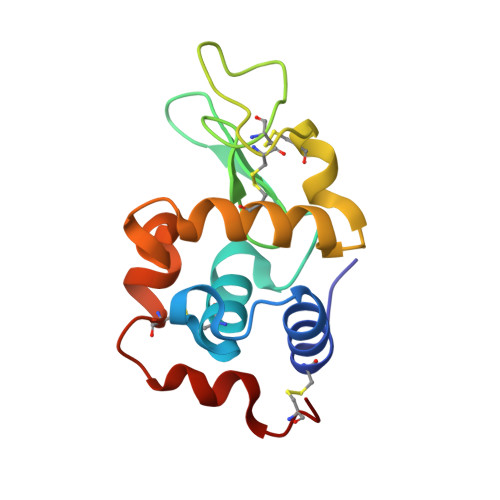
Vanadium compounds have frequently been proposed as therapeutics, but their application has been hampered by the lack of information on the different V-containing species that may form and how these interact with blood and cell proteins, and with enzymes. Herein, we report several resolved crystal structures of lysozyme with bound V IV O 2+ and V IV OL 2+ , where L=2,2'-bipyridine or 1,10-phenanthroline (phen), and of trypsin with V IV O(picolinato) 2 and V V O 2 (phen) + moieties. Computational studies complete the refinement and shed light on the relevant role of hydrophobic interactions, hydrogen bonds, and microsolvation in stabilizating the structure. Noteworthy is that the trypsin-V V O 2 (phen) and trypsin-V IV O(OH)(phen) adducts correspond to similar energies, thus suggesting a possible interconversion under physiological/biological conditions. The obtained data support the relevance of hydrolysis of V IV and V V complexes in the several types of binding established with proteins and the formation of different adducts that might contribute to their pharmacological action, and significantly widen our knowledge of vanadium-protein interactions.
Organizational Affiliation:
Centro de Química Estrutural and Departamento de Engenharia Química, Institute of Molecular Sciences, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001, Lisboa, Portugal.