Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II.
Leenders, R.G.G., Brinch, S.A., Sowa, S.T., Amundsen-Isaksen, E., Galera-Prat, A., Murthy, S., Aertssen, S., Smits, J.N., Nieczypor, P., Damen, E., Wegert, A., Nazare, M., Lehtio, L., Waaler, J., Krauss, S.(2021) J Med Chem 64: 17936-17949
- PubMed: 34878777
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01264
- Primary Citation of Related Structures:
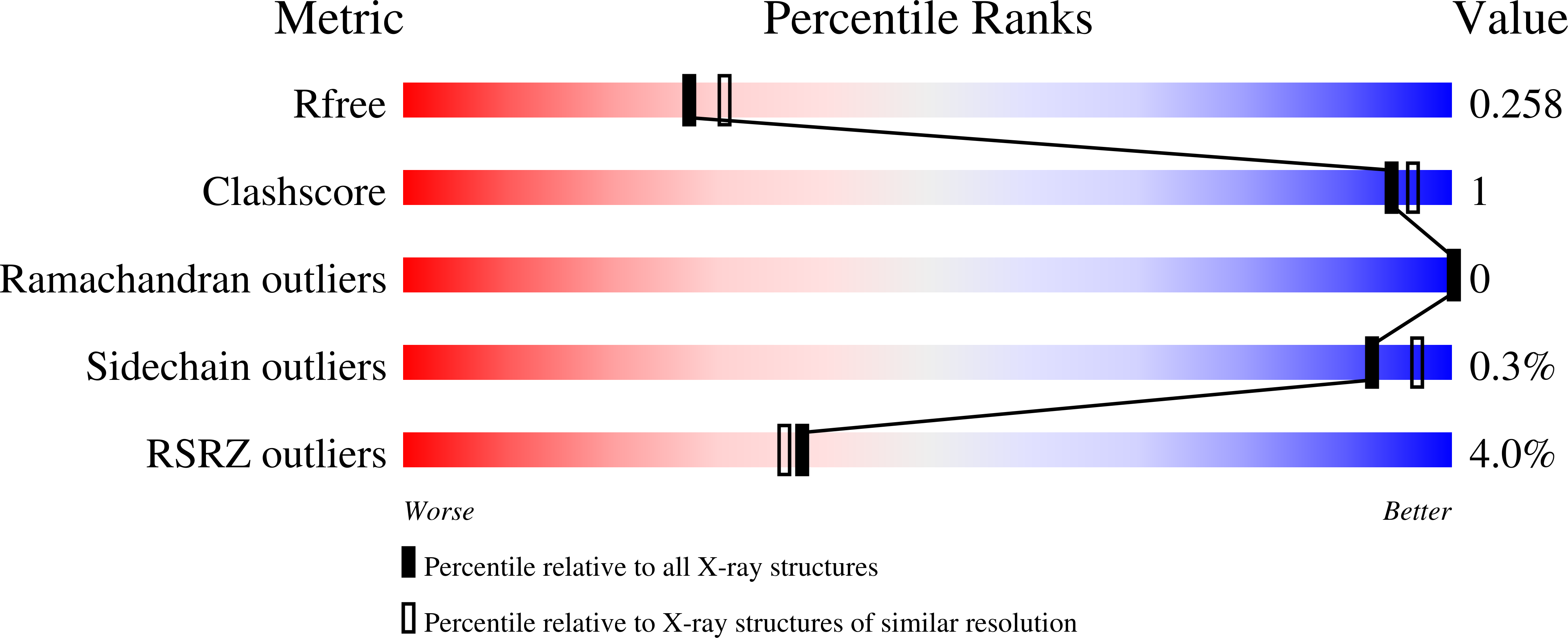
7O6X - PubMed Abstract:
Tankyrase 1 and 2 (TNKS1/2) catalyze post-translational modification by poly-ADP-ribosylation of a plethora of target proteins. In this function, TNKS1/2 also impact the WNT/β-catenin and Hippo signaling pathways that are involved in numerous human disease conditions including cancer. Targeting TNKS1/2 with small-molecule inhibitors shows promising potential to modulate the involved pathways, thereby potentiating disease intervention. Based on our 1,2,4-triazole-based lead compound 1 (OM-1700), further structure-activity relationship analyses of East-, South- and West-single-point alterations and hybrids identified compound 24 (OM-153). Compound 24 showed picomolar IC 50 inhibition in a cellular (HEK293) WNT/β-catenin signaling reporter assay, no off-target liabilities, overall favorable absorption, distribution, metabolism, and excretion (ADME) properties, and an improved pharmacokinetic profile in mice. Moreover, treatment with compound 24 induced dose-dependent biomarker engagement and reduced cell growth in the colon cancer cell line COLO 320DM.
Organizational Affiliation:
Symeres, Kerkenbos 1013, 6546 BB Nijmegen, The Netherlands.