Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Farnung, L., Ochmann, M., Engeholm, M., Cramer, P.(2021) Nat Struct Mol Biol 28: 382-387
- PubMed: 33846633
- DOI: https://doi.org/10.1038/s41594-021-00578-6
- Primary Citation of Related Structures:
7NKX, 7NKY - PubMed Abstract:
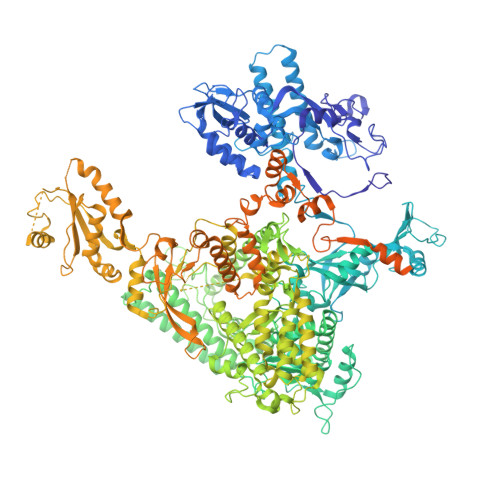
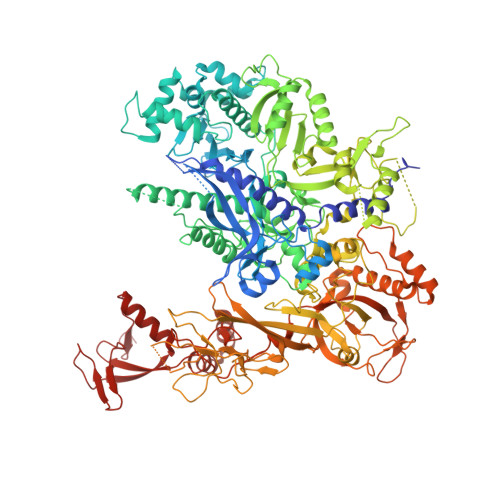
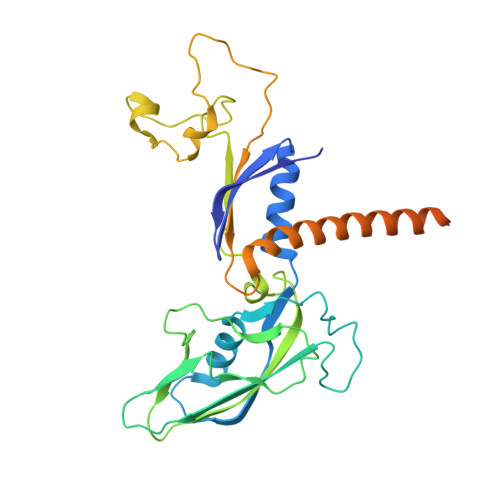
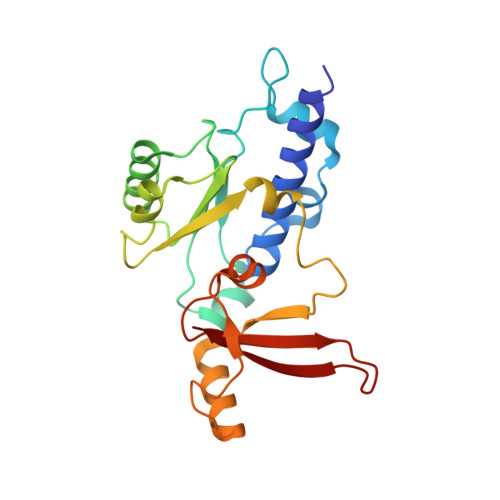
Efficient transcription of RNA polymerase II (Pol II) through nucleosomes requires the help of various factors. Here we show biochemically that Pol II transcription through a nucleosome is facilitated by the chromatin remodeler Chd1 and the histone chaperone FACT when the elongation factors Spt4/5 and TFIIS are present. We report cryo-EM structures of transcribing Saccharomyces cerevisiae Pol II-Spt4/5-nucleosome complexes with bound Chd1 or FACT. In the first structure, Pol II transcription exposes the proximal histone H2A-H2B dimer that is bound by Spt5. Pol II has also released the inhibitory DNA-binding region of Chd1 that is poised to pump DNA toward Pol II. In the second structure, Pol II has generated a partially unraveled nucleosome that binds FACT, which excludes Chd1 and Spt5. These results suggest that Pol II progression through a nucleosome activates Chd1, enables FACT binding and eventually triggers transfer of FACT together with histones to upstream DNA.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.
Organizational Affiliation: