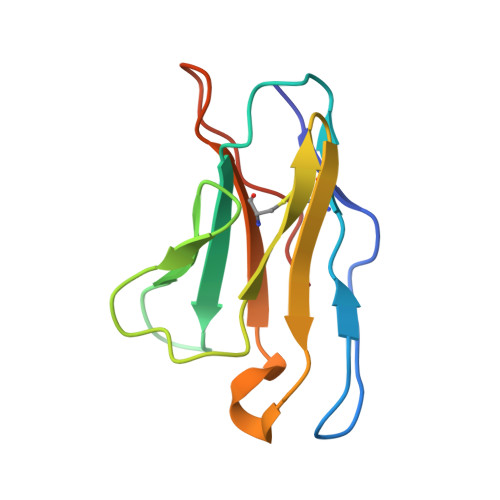
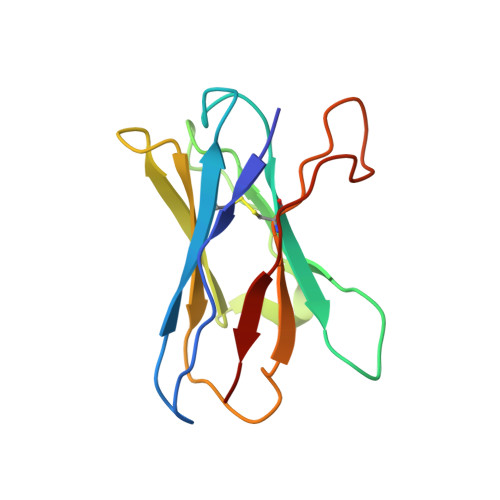
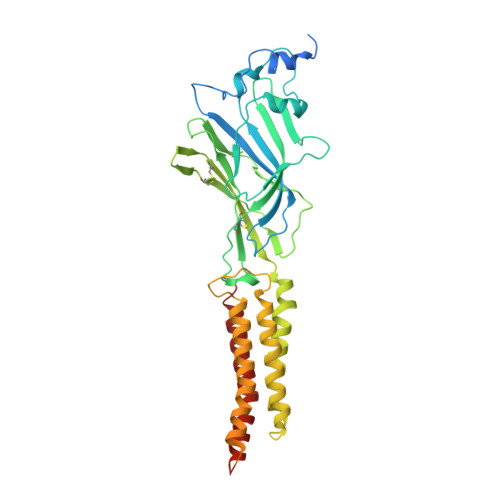
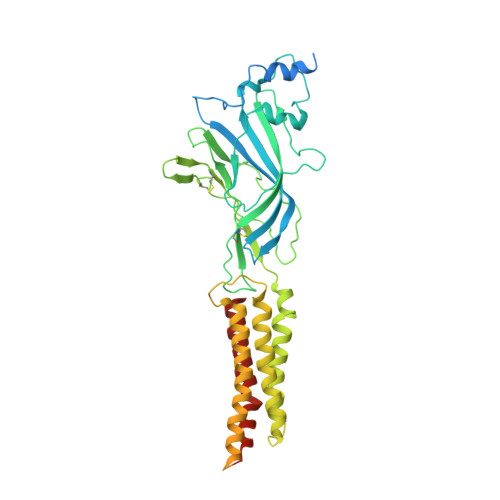
Architecture and assembly mechanism of native glycine receptors.
Zhu, H., Gouaux, E.(2021) Nature 599: 513-517
- PubMed: 34555840
- DOI: https://doi.org/10.1038/s41586-021-04022-z
- Primary Citation of Related Structures:
7MLU, 7MLV, 7MLY - PubMed Abstract:
Glycine receptors (GlyRs) are pentameric, 'Cys-loop' receptors that form chloride-permeable channels and mediate fast inhibitory signalling throughout the central nervous system 1,2 . In the spinal cord and brainstem, GlyRs regulate locomotion and cause movement disorders when mutated 2,3 . However, the stoichiometry of native GlyRs and the mechanism by which they are assembled remain unclear, despite extensive investigation 4-8 . Here we report cryo-electron microscopy structures of native GlyRs from pig spinal cord and brainstem, revealing structural insights into heteromeric receptors and their predominant subunit stoichiometry of 4α:1β. Within the heteromeric pentamer, the β(+)-α(-) interface adopts a structure that is distinct from the α(+)-α(-) and α(+)-β(-) interfaces. Furthermore, the β-subunit contains a unique phenylalanine residue that resides within the pore and disrupts the canonical picrotoxin site. These results explain why inclusion of the β-subunit breaks receptor symmetry and alters ion channel pharmacology. We also find incomplete receptor complexes and, by elucidating their structures, reveal the architectures of partially assembled α-trimers and α-tetramers.
- Vollum Institute, Oregon Health and Science University, Portland, OR, USA.
Organizational Affiliation: