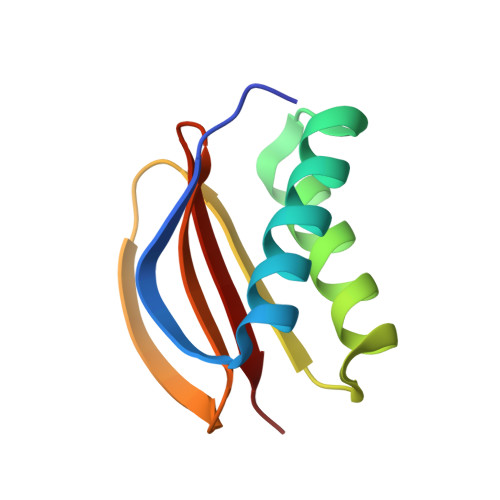
Molecular principles of Piwi-mediated cotranscriptional silencing through the dimeric SFiNX complex.
Schnabl, J., Wang, J., Hohmann, U., Gehre, M., Batki, J., Andreev, V.I., Purkhauser, K., Fasching, N., Duchek, P., Novatchkova, M., Mechtler, K., Plaschka, C., Patel, D.J., Brennecke, J.(2021) Genes Dev 35: 392-409
- PubMed: 33574069
- DOI: https://doi.org/10.1101/gad.347989.120
- Primary Citation of Related Structures:
7K3J, 7K3K, 7K3L - PubMed Abstract:
Nuclear Argonaute proteins, guided by their bound small RNAs to nascent target transcripts, mediate cotranscriptional silencing of transposons and repetitive genomic loci through heterochromatin formation. The molecular mechanisms involved in this process are incompletely understood. Here, we show that the SFiNX complex, a silencing mediator downstream from nuclear Piwi-piRNA complexes in Drosophila , facilitates cotranscriptional silencing as a homodimer. The dynein light chain protein Cut up/LC8 mediates SFiNX dimerization, and its function can be bypassed by a heterologous dimerization domain, arguing for a constitutive SFiNX dimer. Dimeric, but not monomeric SFiNX, is capable of forming molecular condensates in a nucleic acid-stimulated manner. Mutations that prevent SFiNX dimerization result in loss of condensate formation in vitro and the inability of Piwi to initiate heterochromatin formation and silence transposons in vivo. We propose that multivalent SFiNX-nucleic acid interactions are critical for heterochromatin establishment at piRNA target loci in a cotranscriptional manner.
- Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter (VBC), 1030 Vienna, Austria.
Organizational Affiliation: