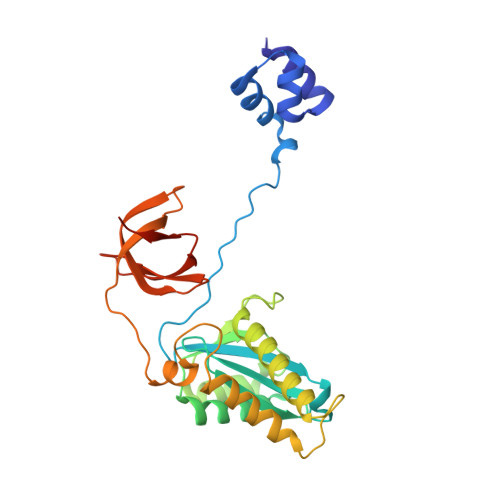
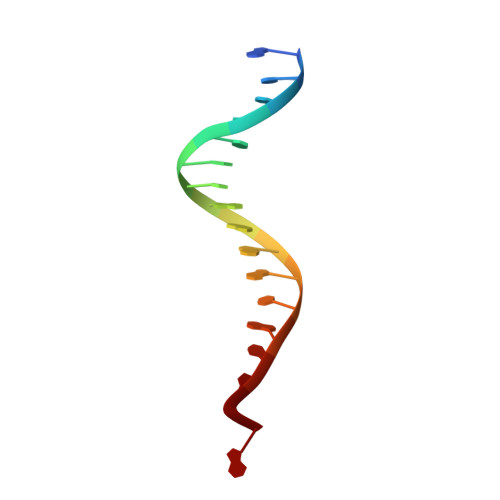
Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Pandey, K.K., Bera, S., Shi, K., Rau, M.J., Oleru, A.V., Fitzpatrick, J.A.J., Engelman, A.N., Aihara, H., Grandgenett, D.P.(2021) Commun Biol 4: 330-330
- PubMed: 33712691
- DOI: https://doi.org/10.1038/s42003-021-01855-2
- Primary Citation of Related Structures:
7JN3, 7KU7, 7KUI - PubMed Abstract:
Despite conserved catalytic integration mechanisms, retroviral intasomes composed of integrase (IN) and viral DNA possess diverse structures with variable numbers of IN subunits. To investigate intasome assembly mechanisms, we employed the Rous sarcoma virus (RSV) IN dimer that assembles a precursor tetrameric structure in transit to the mature octameric intasome. We determined the structure of RSV octameric intasome stabilized by a HIV-1 IN strand transfer inhibitor using single particle cryo-electron microscopy. The structure revealed significant flexibility of the two non-catalytic distal IN dimers along with previously unrecognized movement of the conserved intasome core, suggesting ordered conformational transitions between intermediates that may be important to capture the target DNA. Single amino acid substitutions within the IN C-terminal domain affected intasome assembly and function in vitro and infectivity of pseudotyped RSV virions. Unexpectedly, 17 C-terminal amino acids of IN were dispensable for virus infection despite regulating the transition of the tetrameric intasome to the octameric form in vitro. We speculate that this region may regulate the binding of highly flexible distal IN dimers to the intasome core to form the octameric complex. Our studies reveal key steps in the assembly of RSV intasomes.
- Department of Molecular Microbiology and Immunology, School of Medicine, Saint Louis University, St. Louis, MO, USA. krishan.pandey@health.slu.edu.
Organizational Affiliation: