Structure of the human CLCN7-OSTM1 complex with ATP
Yang, G.H., Lu, Y.M., Zhang, Y.M., Jia, Y.T., Li, X.M., Lei, J.L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Osteopetrosis-associated transmembrane protein 1 | 344 | Homo sapiens | Mutation(s): 0 Gene Names: OSTM1, GL, HSPC019, UNQ6098/PRO21201 Membrane Entity: Yes | 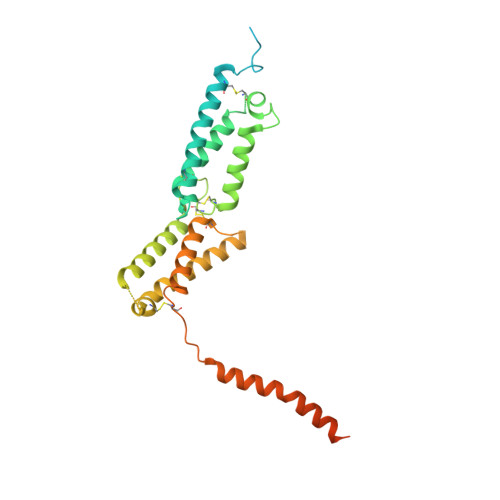 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q86WC4 (Homo sapiens) Explore Q86WC4 Go to UniProtKB: Q86WC4 | |||||
PHAROS: Q86WC4 GTEx: ENSG00000081087 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86WC4 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: Q86WC4-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| H(+)/Cl(-) exchange transporter 7 | 825 | Homo sapiens | Mutation(s): 0 Gene Names: CLCN7 Membrane Entity: Yes | 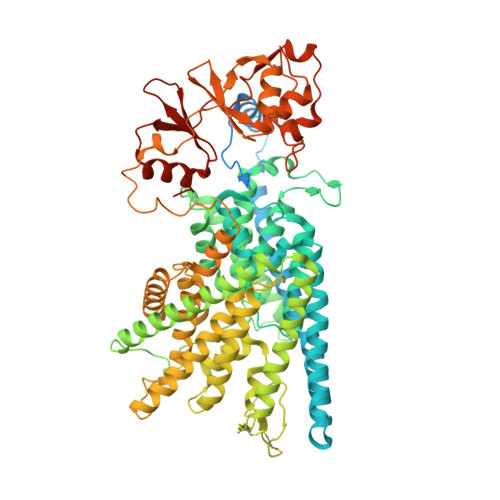 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P51798 (Homo sapiens) Explore P51798 Go to UniProtKB: P51798 | |||||
PHAROS: P51798 GTEx: ENSG00000103249 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P51798 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | G [auth C], J [auth D] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| CL Query on CL | H [auth C], I [auth C], K [auth D], L [auth D] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |