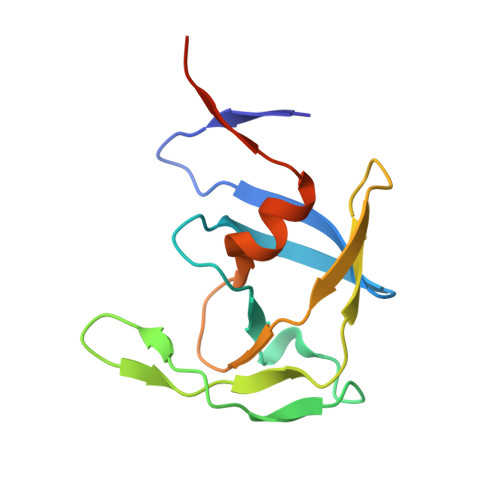
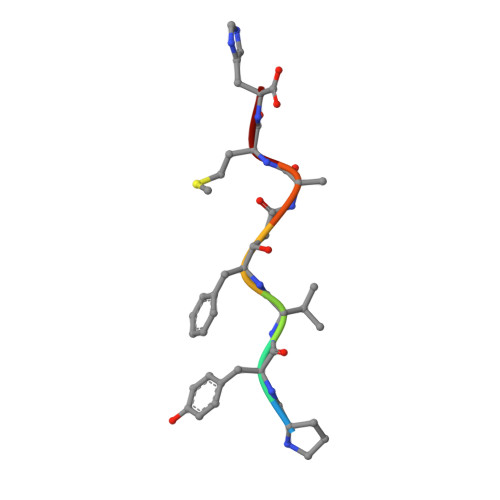
Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Wosicki, S., Kazmierczyk, M., Gilski, M., Zabranska, H., Pichova, I., Jaskolski, M.(2021) Protein Sci 30: 1258-1263
- PubMed: 33786913
- DOI: https://doi.org/10.1002/pro.4072
- Primary Citation of Related Structures:
7BGT, 7BGU - PubMed Abstract:
Mason-Pfizer monkey virus protease (PR) was crystallized in complex with two pepstatin-based inhibitors in P1 space group. In both crystal structures, the extended flap loops that lock the inhibitor/substrate over the active site, are visible in the electron density either completely or with only small gaps, providing the first observation of the conformation of the flap loops in dimeric complex form of this retropepsin. The H-bond network in the active site (with D26N mutation) differs from that reported for the P2 1 crystal structures and is similar to a rarely occurring system in HIV-1 PR.
- Center for Biocrystallographic Research, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.
Organizational Affiliation: