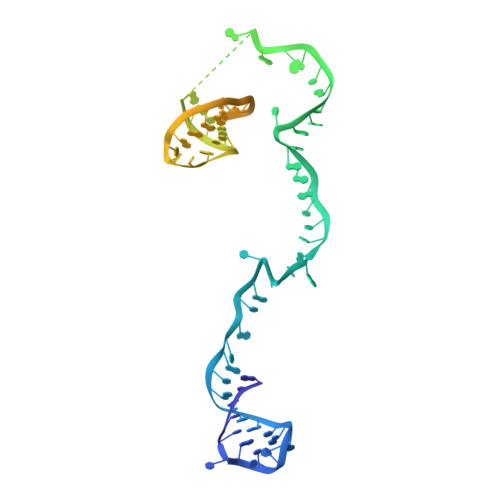
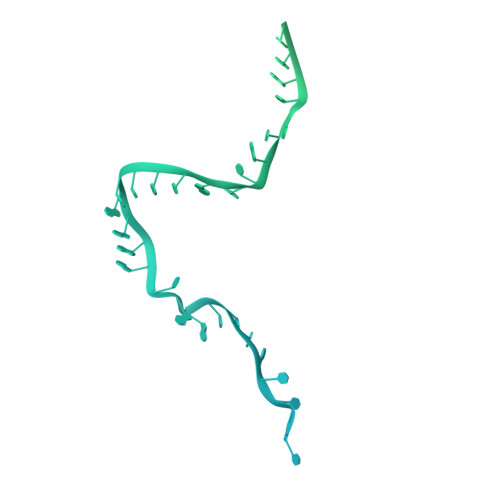
Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Townsend, C., Leelaram, M.N., Agafonov, D.E., Dybkov, O., Will, C.L., Bertram, K., Urlaub, H., Kastner, B., Stark, H., Luhrmann, R.(2020) Science 370
- PubMed: 33243851
- DOI: https://doi.org/10.1126/science.abc3753
- Primary Citation of Related Structures:
7AAV, 7ABF, 7ABG, 7ABH, 7ABI - PubMed Abstract:
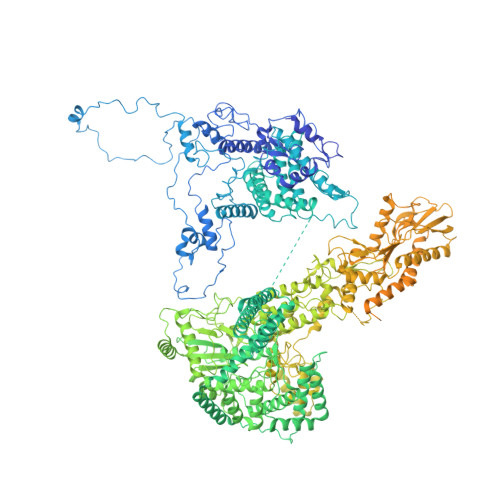
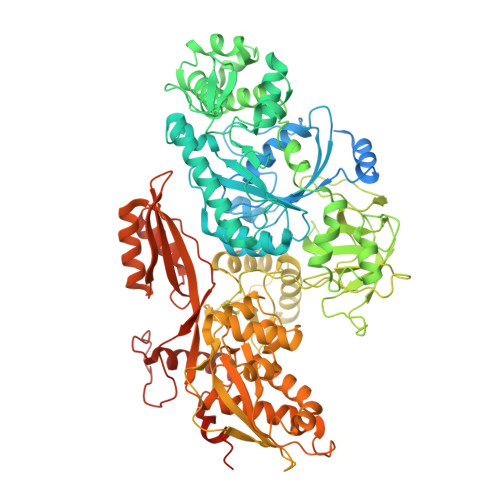
Spliceosome activation involves extensive protein and RNA rearrangements that lead to formation of a catalytically active U2/U6 RNA structure. At present, little is known about the assembly pathway of the latter and the mechanism whereby proteins aid its proper folding. Here, we report the cryo-electron microscopy structures of two human, activated spliceosome precursors (that is, pre-B act complexes) at core resolutions of 3.9 and 4.2 angstroms. These structures elucidate the order of the numerous protein exchanges that occur during activation, the mutually exclusive interactions that ensure the correct order of ribonucleoprotein rearrangements needed to form the U2/U6 catalytic RNA, and the stepwise folding pathway of the latter. Structural comparisons with mature B act complexes reveal the molecular mechanism whereby a conformational change in the scaffold protein PRP8 facilitates final three-dimensional folding of the U2/U6 catalytic RNA.
- Department of Structural Dynamics, MPI for Biophysical Chemistry, Am Fassberg 11, D-37077 Göttingen, Germany.
Organizational Affiliation: