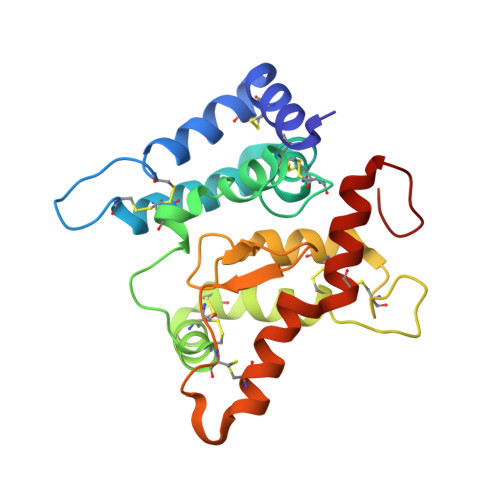
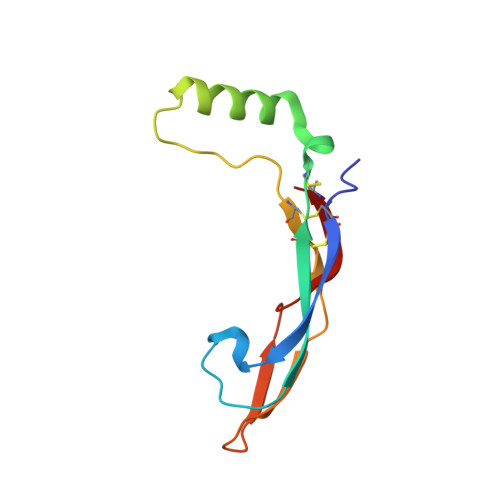
A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Adams, S.E., Purkiss, A.G., Knowles, P.P., Nans, A., Briggs, D.C., Borg, A., Earl, C.P., Goodman, K.M., Nawrotek, A., Borg, A.J., McIntosh, P.B., Houghton, F.M., Kjaer, S., McDonald, N.Q.(2021) Structure 29: 694
- PubMed: 33484636
- DOI: https://doi.org/10.1016/j.str.2020.12.012
- Primary Citation of Related Structures:
7AB8, 7AMK, 7AML - PubMed Abstract:
RET receptor tyrosine kinase plays vital developmental and neuroprotective roles in metazoans. GDNF family ligands (GFLs) when bound to cognate GFRα co-receptors recognize and activate RET stimulating its cytoplasmic kinase function. The principles for RET ligand-co-receptor recognition are incompletely understood. Here, we report a crystal structure of the cadherin-like module (CLD1-4) from zebrafish RET revealing interdomain flexibility between CLD2 and CLD3. Comparison with a cryo-electron microscopy structure of a ligand-engaged zebrafish RET ECD -GDNF-GFRα1a complex indicates conformational changes within a clade-specific CLD3 loop adjacent to the co-receptor. Our observations indicate that RET is a molecular clamp with a flexible calcium-dependent arm that adapts to different GFRα co-receptors, while its rigid arm recognizes a GFL dimer to align both membrane-proximal cysteine-rich domains. We also visualize linear arrays of RET ECD -GDNF-GFRα1a suggesting that a conserved contact stabilizes higher-order species. Our study reveals that ligand-co-receptor recognition by RET involves both receptor plasticity and strict spacing of receptor dimers by GFL ligands.
- Signalling and Structural Biology Laboratory, Francis Crick Institute, NW1 1AT London, UK.
Organizational Affiliation: