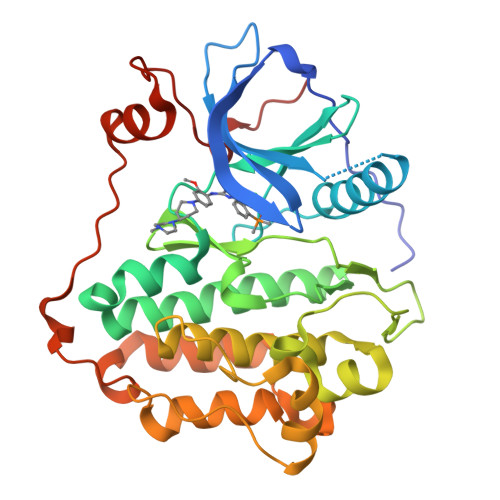
Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Grabe, T., Jeyakumar, K., Niggenaber, J., Schulz, T., Koska, S., Kleinbolting, S., Beck, M.E., Muller, M.P., Rauh, D.(2023) ACS Med Chem Lett 14: 591-598
- PubMed: 37197473
- DOI: https://doi.org/10.1021/acsmedchemlett.2c00514
- Primary Citation of Related Structures:
7ZYM, 7ZYN, 7ZYP, 7ZYQ - PubMed Abstract:
Drug resistance mutations emerging during the treatment of non-small cell lung cancer (NSCLC) with epidermal growth factor receptor (EGFR) inhibitors represent a major challenge in personalized cancer treatment and require constant development of new inhibitors. For the covalent irreversible EGFR inhibitor osimertinib, the predominant resistance mechanism is the acquired C797S mutation, which abolishes the covalent anchor point and thus results in a dramatic loss in potency. In this study, we present next-generation reversible EGFR inhibitors with the potential to overcome this EGFR-C797S resistance mutation. For this, we combined the reversible methylindole-aminopyrimidine scaffold known from osimertinib with the affinity driving isopropyl ester of mobocertinib. By occupying the hydrophobic back pocket, we were able to generate reversible inhibitors with subnanomolar activity against EGFR-L858R/C797S and EGFR-L858R/T790M/C797S with cellular activity on EGFR-L858R/C797S dependent Ba/F3 cells. Additionally, we were able to resolve cocrystal structures of these reversible aminopyrimidines, which will guide further inhibitor design toward C797S-mutated EGFR.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, TU Dortmund University and Drug Discovery Hub Dortmund (DDHD) am Zentrum für Integrierte Wirkstoffforschung (ZIW), Otto-Hahn-Strasse 4a, 44227 Dortmund, Germany.