Large-scale phosphomimetic screening identifies phospho-modulated motif-based protein interactions.
Kliche, J., Garvanska, D.H., Simonetti, L., Badgujar, D., Dobritzsch, D., Nilsson, J., Davey, N.E., Ivarsson, Y.(2023) Mol Syst Biol 19: e11164-e11164
- PubMed: 37219487
- DOI: https://doi.org/10.15252/msb.202211164
- Primary Citation of Related Structures:
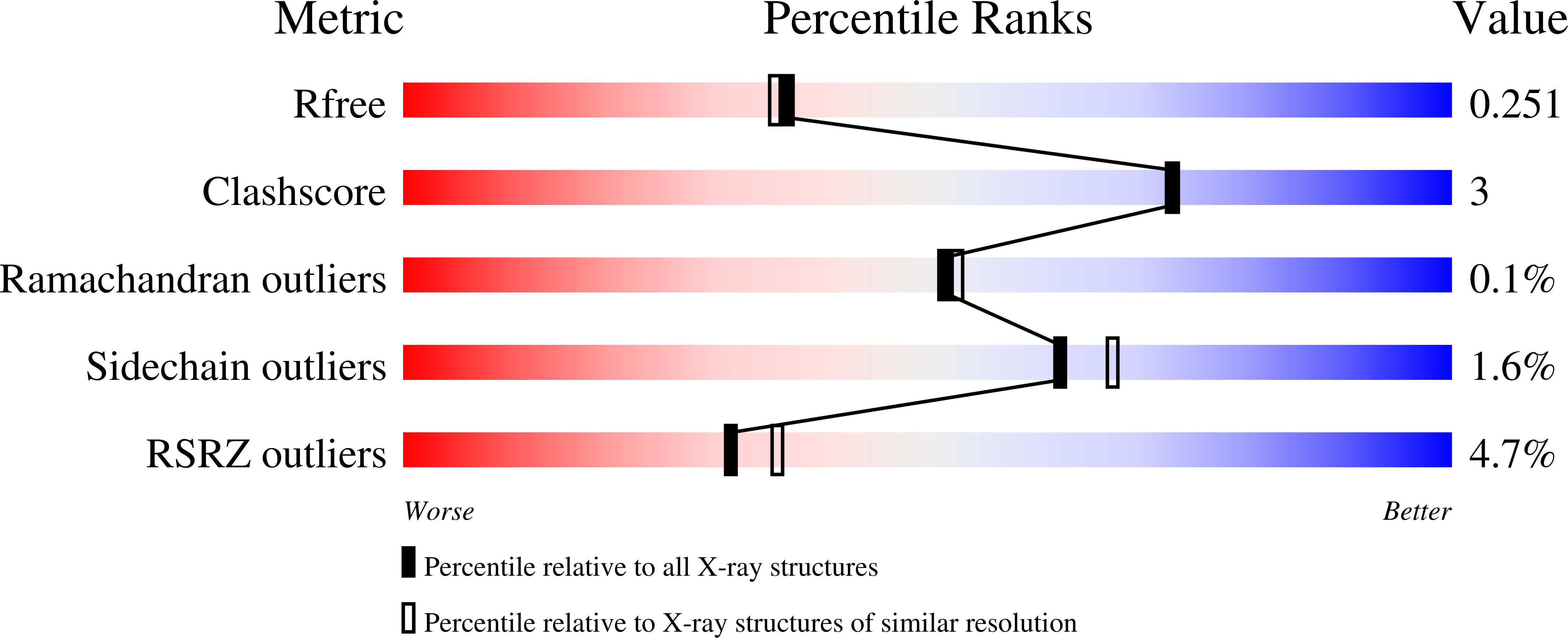
7ZX4 - PubMed Abstract:
Phosphorylation is a ubiquitous post-translation modification that regulates protein function by promoting, inhibiting or modulating protein-protein interactions. Hundreds of thousands of phosphosites have been identified but the vast majority have not been functionally characterised and it remains a challenge to decipher phosphorylation events modulating interactions. We generated a phosphomimetic proteomic peptide-phage display library to screen for phosphosites that modulate short linear motif-based interactions. The peptidome covers ~13,500 phospho-serine/threonine sites found in the intrinsically disordered regions of the human proteome. Each phosphosite is represented as wild-type and phosphomimetic variant. We screened 71 protein domains to identify 248 phosphosites that modulate motif-mediated interactions. Affinity measurements confirmed the phospho-modulation of 14 out of 18 tested interactions. We performed a detailed follow-up on a phospho-dependent interaction between clathrin and the mitotic spindle protein hepatoma-upregulated protein (HURP), demonstrating the essentiality of the phospho-dependency to the mitotic function of HURP. Structural characterisation of the clathrin-HURP complex elucidated the molecular basis for the phospho-dependency. Our work showcases the power of phosphomimetic ProP-PD to discover novel phospho-modulated interactions required for cellular function.
Organizational Affiliation:
Department of Chemistry, BMC, Uppsala University, Uppsala, Sweden.