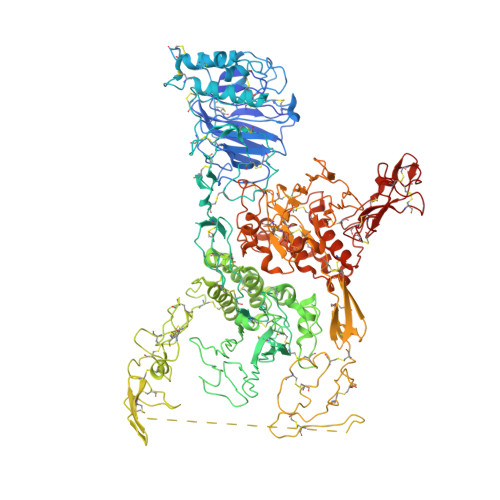
Assembly of von Willebrand factor tubules with in vivo helical parameters requires A1 domain insertion.
Javitt, G., Yeshaya, N., Khmelnitsky, L., Fass, D.(2022) Blood 140: 2835-2843
- PubMed: 36179246
- DOI: https://doi.org/10.1182/blood.2022017153
- Primary Citation of Related Structures:
7ZWH - PubMed Abstract:
The von Willebrand factor (VWF) glycoprotein is stored in tubular form in Weibel-Palade bodies (WPBs) before secretion from endothelial cells into the bloodstream. The organization of VWF in the tubules promotes formation of covalently linked VWF polymers and enables orderly secretion without polymer tangling. Recent studies have described the high-resolution structure of helical tubular cores formed in vitro by the D1D2 and D'D3 amino-terminal protein segments of VWF. Here we show that formation of tubules with the helical geometry observed for VWF in intracellular WPBs requires also the VWA1 (A1) domain. We reconstituted VWF tubules from segments containing the A1 domain and discovered it to be inserted between helical turns of the tubule, altering helical parameters and explaining the increased robustness of tubule formation when A1 is present. The conclusion from this observation is that the A1 domain has a direct role in VWF assembly, along with its known activity in hemostasis after secretion.
Organizational Affiliation:
Department of Chemical and Structural Biology, Weizmann Institute of Science, Rehovot, Israel.