Biochemical, structural, and computational studies of a gamma-carbonic anhydrase from the pathogenic bacterium Burkholderia pseudomallei.
Di Fiore, A., De Luca, V., Langella, E., Nocentini, A., Buonanno, M., Maria Monti, S., Supuran, C.T., Capasso, C., De Simone, G.(2022) Comput Struct Biotechnol J 20: 4185-4194
- PubMed: 36016712
- DOI: https://doi.org/10.1016/j.csbj.2022.07.033
- Primary Citation of Related Structures:
7ZW9 - PubMed Abstract:
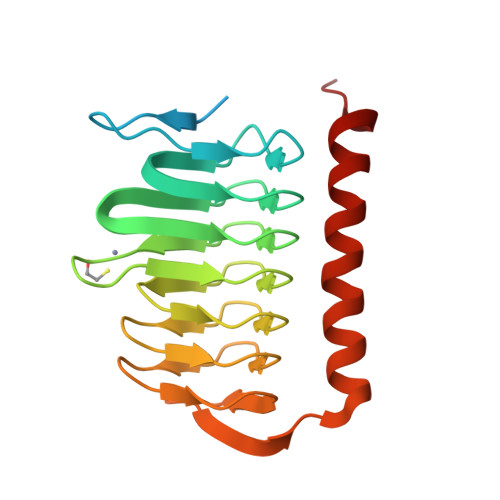
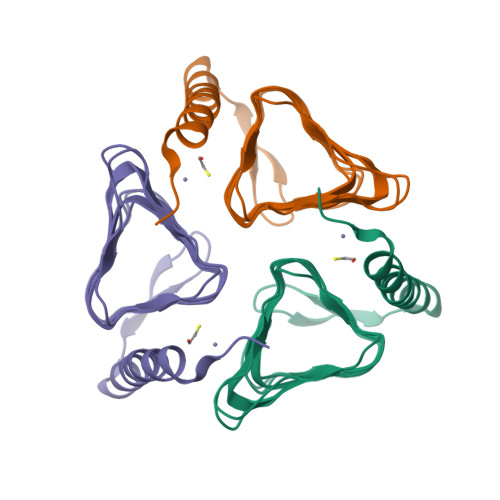
Melioidosis is a severe disease caused by the highly pathogenic gram-negative bacterium Burkholderia pseudomallei . Several studies have highlighted the broad resistance of this pathogen to many antibiotics and pointed out the pivotal importance of improving the pharmacological arsenal against it. Since γ-carbonic anhydrases (γ-CAs) have been recently introduced as potential and novel antibacterial drug targets, in this paper, we report a detailed characterization of BpsγCA, a γ-CA from B . pseudomallei by a multidisciplinary approach. In particular, the enzyme was recombinantly produced and biochemically characterized. Its catalytic activity at different pH values was measured, the crystal structure was determined and theoretical pKa calculations were carried out. Results provided a snapshot of the enzyme active site and dissected the role of residues involved in the catalytic mechanism and ligand recognition. These findings are an important starting point for developing new anti-melioidosis drugs targeting BpsγCA.
Organizational Affiliation:
Institute of Biostructures and Bioimaging-CNR, via Pietro Castellino 111, 80131 Napoli, Italy.