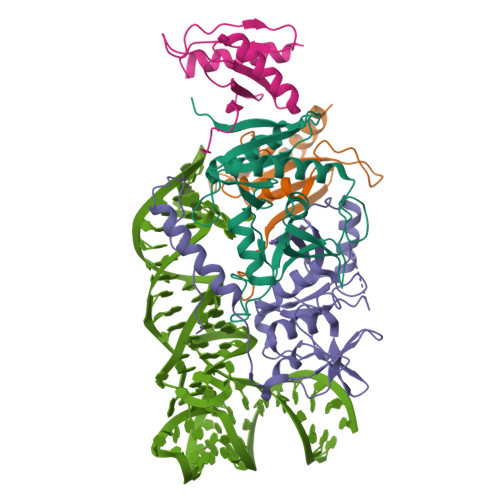
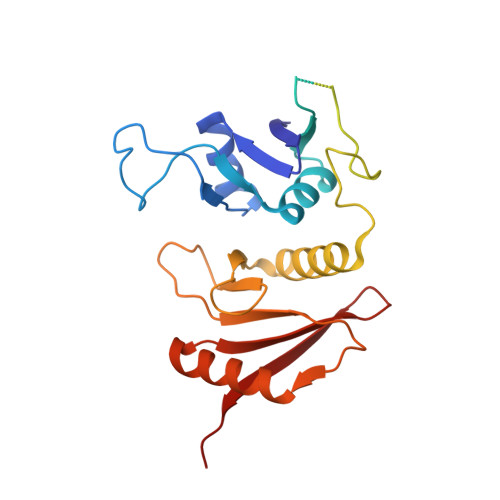
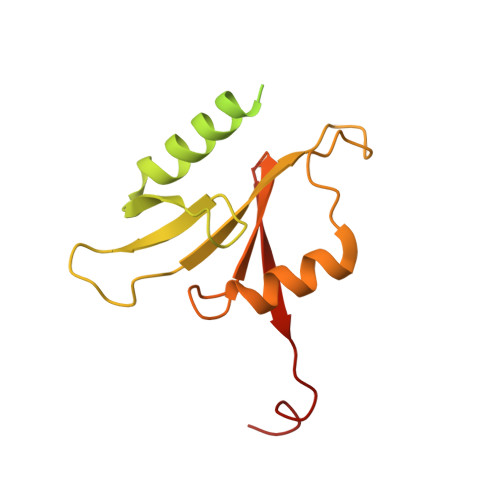
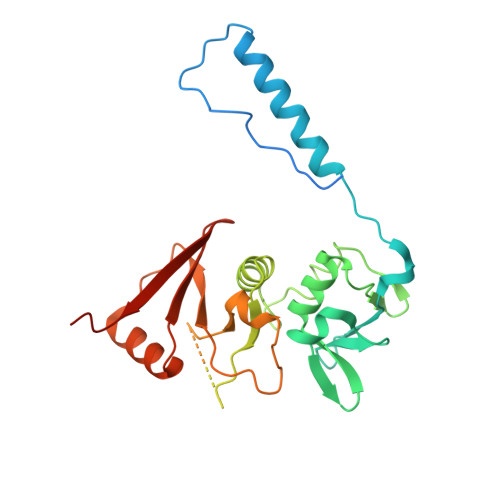
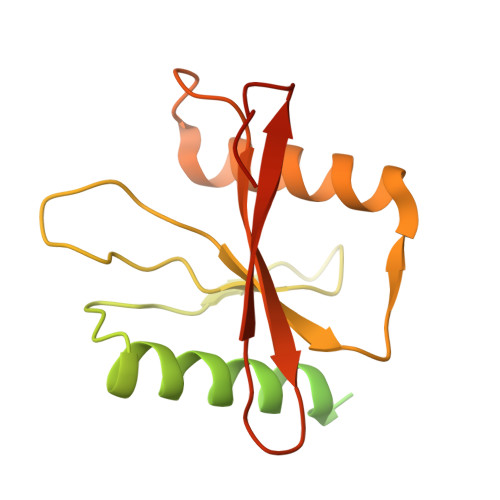
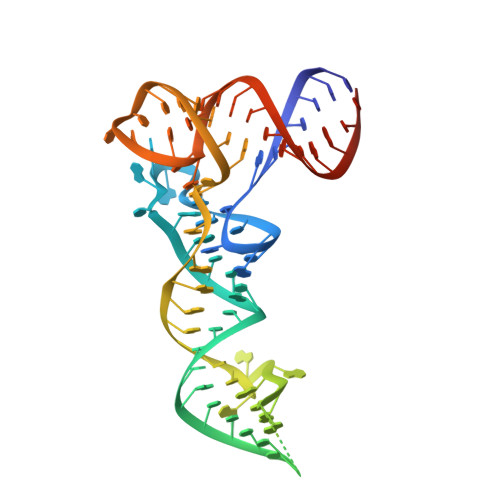
Structural basis of substrate recognition by human tRNA splicing endonuclease TSEN.
Sekulovski, S., Susac, L., Stelzl, L.S., Tampe, R., Trowitzsch, S.(2023) Nat Struct Mol Biol 30: 834-840
- PubMed: 37231152
- DOI: https://doi.org/10.1038/s41594-023-00992-y
- Primary Citation of Related Structures:
7ZRZ - PubMed Abstract:
Heterotetrameric human transfer RNA (tRNA) splicing endonuclease TSEN catalyzes intron excision from precursor tRNAs (pre-tRNAs), utilizing two composite active sites. Mutations in TSEN and its associated RNA kinase CLP1 are linked to the neurodegenerative disease pontocerebellar hypoplasia (PCH). Despite the essential function of TSEN, the three-dimensional assembly of TSEN-CLP1, the mechanism of substrate recognition, and the structural consequences of disease mutations are not understood in molecular detail. Here, we present single-particle cryogenic electron microscopy reconstructions of human TSEN with intron-containing pre-tRNAs. TSEN recognizes the body of pre-tRNAs and pre-positions the 3' splice site for cleavage by an intricate protein-RNA interaction network. TSEN subunits exhibit large unstructured regions flexibly tethering CLP1. Disease mutations localize far from the substrate-binding interface and destabilize TSEN. Our work delineates molecular principles of pre-tRNA recognition and cleavage by human TSEN and rationalizes mutations associated with PCH.
Organizational Affiliation:
Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Frankfurt am Main, Germany.