Mechanisms of inward transmembrane proton translocation.
Kovalev, K., Tsybrov, F., Alekseev, A., Shevchenko, V., Soloviov, D., Siletsky, S., Bourenkov, G., Agthe, M., Nikolova, M., von Stetten, D., Astashkin, R., Bukhdruker, S., Chizhov, I., Royant, A., Kuzmin, A., Gushchin, I., Rosselli, R., Rodriguez-Valera, F., Ilyinskiy, N., Rogachev, A., Borshchevskiy, V., Schneider, T.R., Bamberg, E., Gordeliy, V.(2023) Nat Struct Mol Biol 30: 970-979
- PubMed: 37386213
- DOI: https://doi.org/10.1038/s41594-023-01020-9
- Primary Citation of Related Structures:
7ZMY, 7ZN0, 7ZN3, 7ZN8, 7ZN9, 7ZNA, 7ZNB, 7ZNC, 7ZND, 7ZNE, 7ZNG, 7ZNH, 7ZNI - PubMed Abstract:
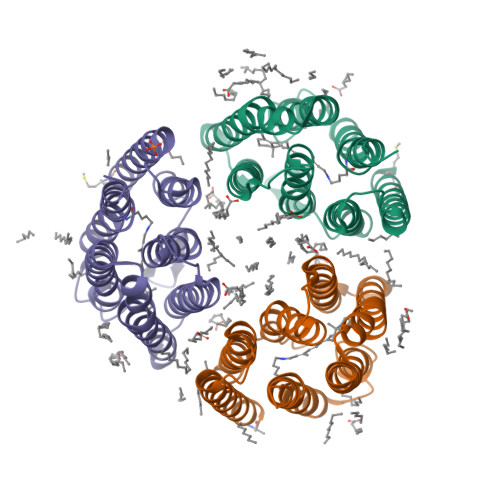
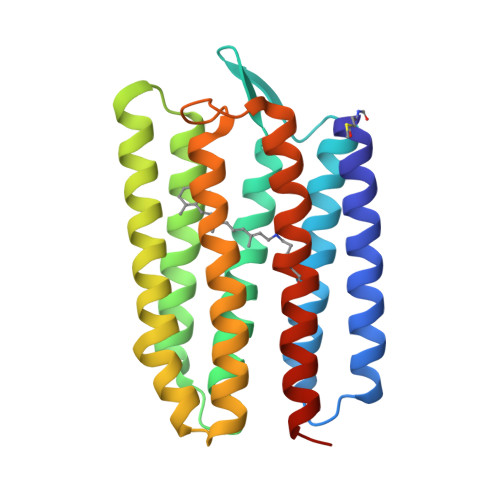
Proton transport is indispensable for cell life. It is believed that molecular mechanisms of proton movement through different types of proton-conducting molecules have general universal features. However, elucidation of such mechanisms is a challenge. It requires true-atomic-resolution structures of all key proton-conducting states. Here we present a comprehensive function-structure study of a light-driven bacterial inward proton pump, xenorhodopsin, from Bacillus coahuilensis in all major proton-conducting states. The structures reveal that proton translocation is based on proton wires regulated by internal gates. The wires serve as both selectivity filters and translocation pathways for protons. The cumulative results suggest a general concept of proton translocation. We demonstrate the use of serial time-resolved crystallography at a synchrotron source with sub-millisecond resolution for rhodopsin studies, opening the door for principally new applications. The results might also be of interest for optogenetics since xenorhodopsins are the only alternative tools to fire neurons.
Organizational Affiliation:
European Molecular Biology Laboratory, Hamburg unit c/o DESY, Hamburg, Germany.