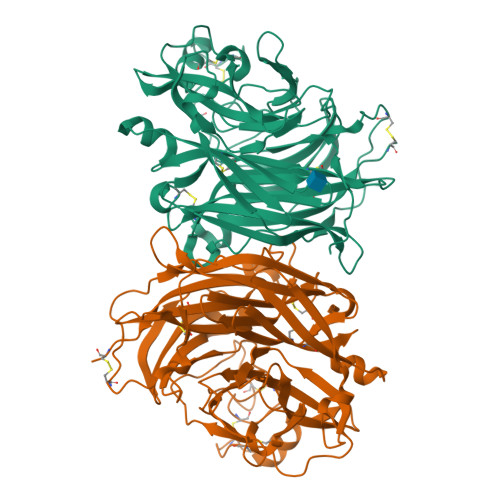
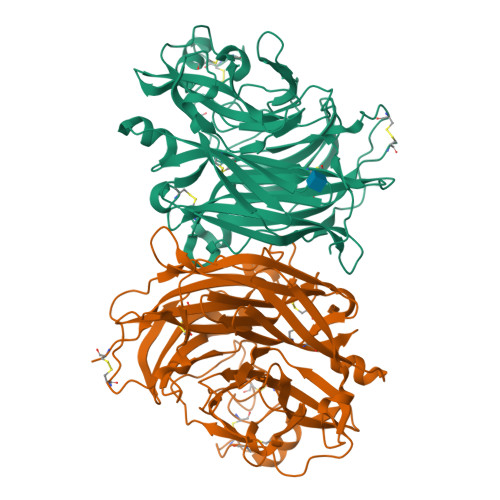
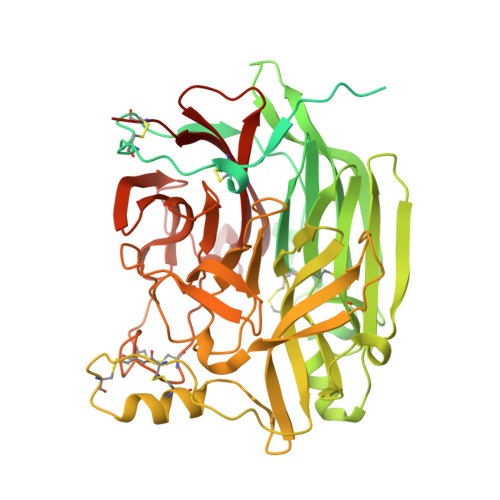
Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Stelfox, A.J., Oguntuyo, K.Y., Rissanen, I., Harlos, K., Rambo, R., Lee, B., Bowden, T.A.(2023) mBio 14: e0139123-e0139123
- PubMed: 37737607
- DOI: https://doi.org/10.1128/mbio.01391-23
- Primary Citation of Related Structures:
7ZM5, 7ZM6 - PubMed Abstract:
Genetically diverse paramyxoviruses are united in their presentation of a receptor-binding protein (RBP), which works in concert with the fusion protein to facilitate host-cell entry. The C-terminal head region of the paramyxoviral RBP, a primary determinant of host-cell tropism and inter-species transmission potential, forms structurally distinct classes dependent upon protein and glycan receptor specificity. Here, we reveal the architecture of the C-terminal head region of the RBPs from Nariva virus (NarV) and Mossman virus (MosV), two archetypal rodent-borne paramyxoviruses within the recently established genus Narmovirus , family Paramyxoviridae . Our analysis reveals that while narmoviruses retain the general architectural features associated with paramyxoviral RBPs, namely, a six-bladed β-propeller fold, they lack the structural motifs associated with known receptor-mediated host-cell entry pathways. This investigation indicates that the RBPs of narmoviruses exhibit pathobiological features that are distinct from those of other paramyxoviruses.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford , Oxford, United Kingdom.