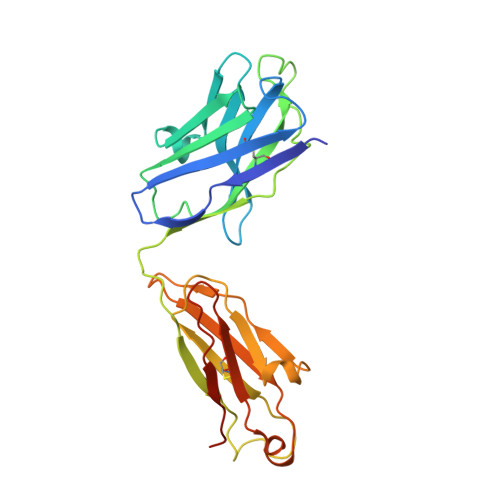
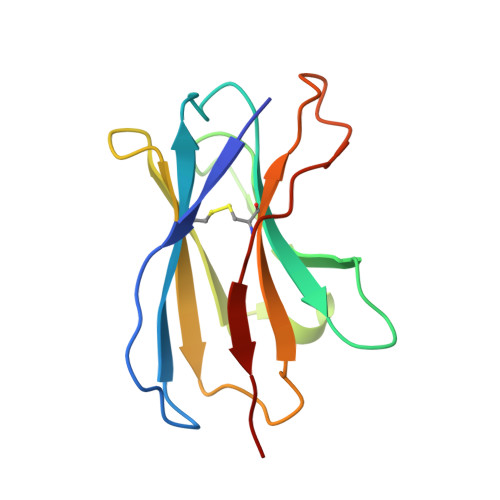
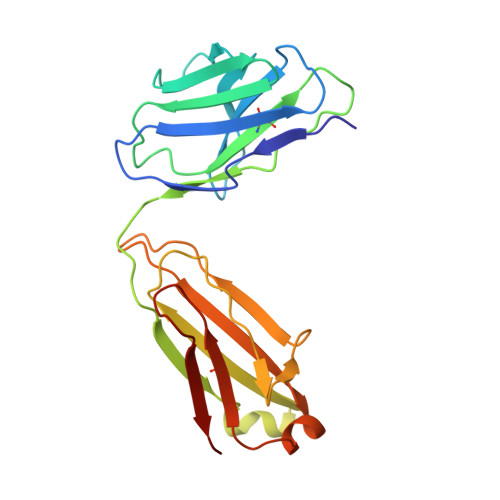
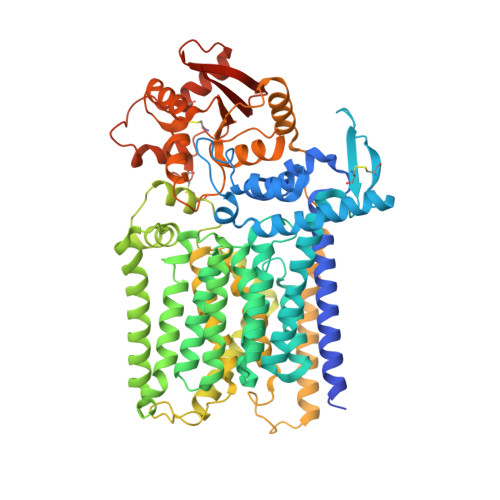
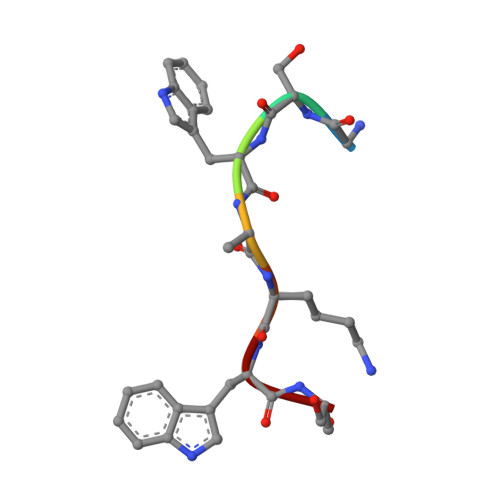
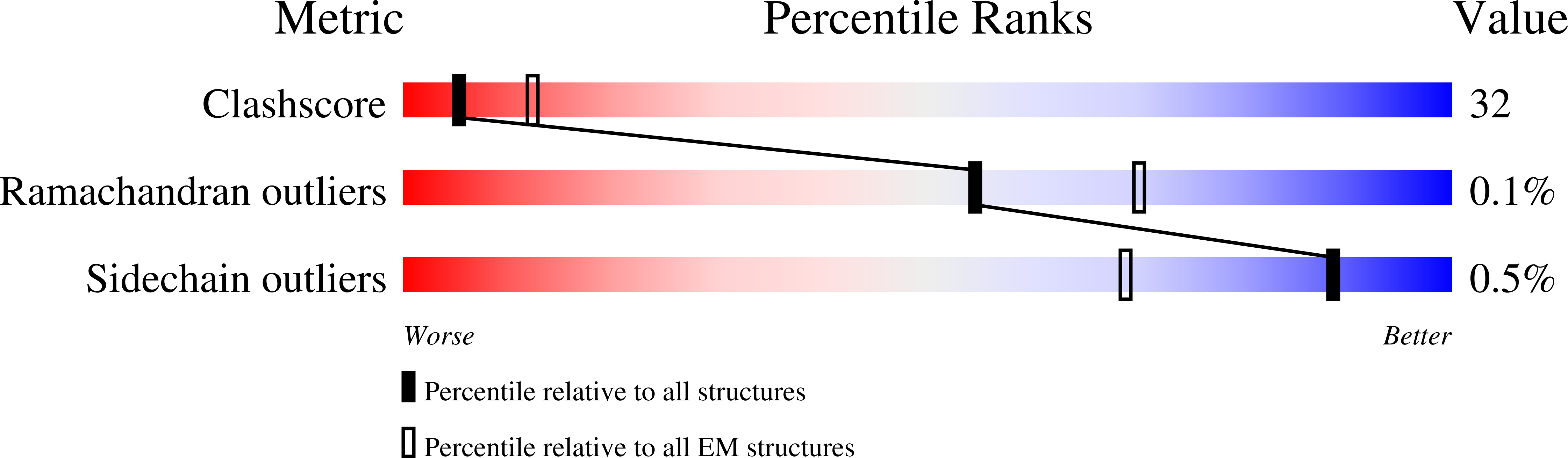Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Bloch, J.S., John, A., Mao, R., Mukherjee, S., Boilevin, J., Irobalieva, R.N., Darbre, T., Scott, N.E., Reymond, J.L., Kossiakoff, A.A., Goddard-Borger, E.D., Locher, K.P.(2023) Nat Chem Biol 19: 575-584
- PubMed: 36604564
- DOI: https://doi.org/10.1038/s41589-022-01219-9
- Primary Citation of Related Structures:
7ZLG, 7ZLH, 7ZLI, 7ZLJ - PubMed Abstract:
C-linked glycosylation is essential for the trafficking, folding and function of secretory and transmembrane proteins involved in cellular communication processes. The tryptophan C-mannosyltransferase (CMT) enzymes that install the modification attach a mannose to the first tryptophan of WxxW/C sequons in nascent polypeptide chains by an unknown mechanism. Here, we report cryogenic-electron microscopy structures of Caenorhabditis elegans CMT in four key states: apo, acceptor peptide-bound, donor-substrate analog-bound and as a trapped ternary complex with both peptide and a donor-substrate mimic bound. The structures indicate how the C-mannosylation sequon is recognized by this CMT and its paralogs, and how sequon binding triggers conformational activation of the donor substrate: a process relevant to all glycosyltransferase C superfamily enzymes. Our structural data further indicate that the CMTs adopt an unprecedented electrophilic aromatic substitution mechanism to enable the C-glycosylation of proteins. These results afford opportunities for understanding human disease and therapeutic targeting of specific CMT paralogs.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, Zürich, Switzerland.