Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Marcinkiewicz, A.L., Brangulis, K., Dupuis 2nd, A.P., Hart, T.M., Zamba-Campero, M., Nowak, T.A., Stout, J.L., Akopjana, I., Kazaks, A., Bogans, J., Ciota, A.T., Kraiczy, P., Kolokotronis, S.O., Lin, Y.P.(2023) Proc Natl Acad Sci U S A 120: e2301549120-e2301549120
- PubMed: 37364114
- DOI: https://doi.org/10.1073/pnas.2301549120
- Primary Citation of Related Structures:
7ZJJ, 7ZJK, 7ZJM - PubMed Abstract:
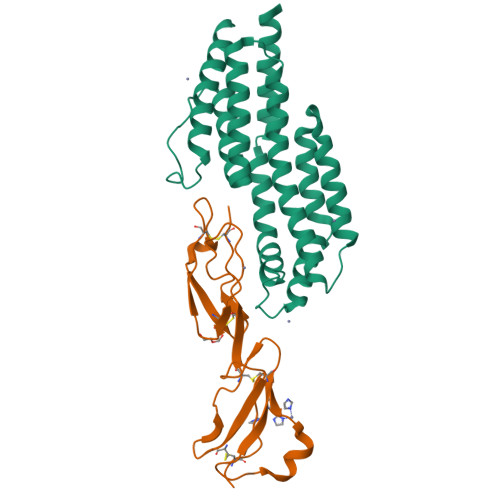
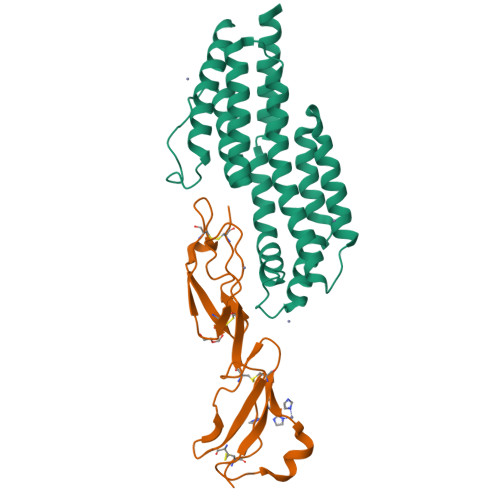
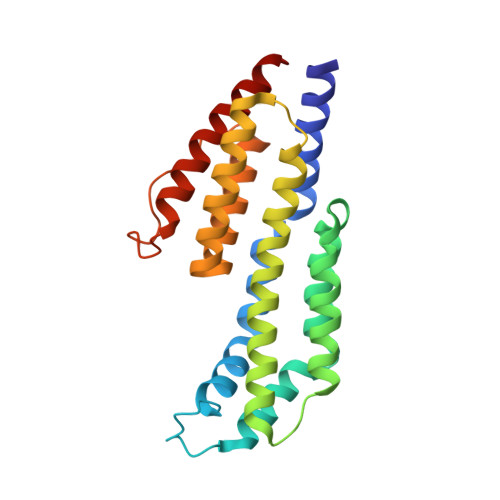
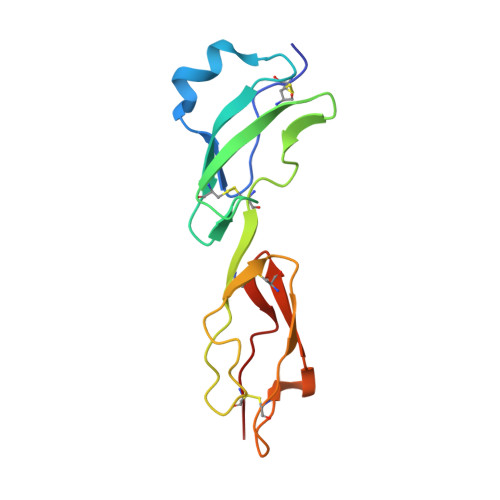
Modern infectious disease outbreaks often involve changes in host tropism, the preferential adaptation of pathogens to specific hosts. The Lyme disease-causing bacterium Borrelia burgdorferi ( Bb ) is an ideal model to investigate the molecular mechanisms of host tropism, because different variants of these tick-transmitted bacteria are distinctly maintained in rodents or bird reservoir hosts. To survive in hosts and escape complement-mediated immune clearance, Bb produces the outer surface protein CspZ that binds the complement inhibitor factor H (FH) to facilitate bacterial dissemination in vertebrates. Despite high sequence conservation, CspZ variants differ in human FH-binding ability. Together with the FH polymorphisms between vertebrate hosts, these findings suggest that minor sequence variation in this bacterial outer surface protein may confer dramatic differences in host-specific, FH-binding-mediated infectivity. We tested this hypothesis by determining the crystal structure of the CspZ-human FH complex, and identifying minor variation localized in the FH-binding interface yielding bird and rodent FH-specific binding activity that impacts infectivity. Swapping the divergent region in the FH-binding interface between rodent- and bird-associated CspZ variants alters the ability to promote rodent- and bird-specific early-onset dissemination. We further linked these loops and respective host-specific, complement-dependent phenotypes with distinct CspZ phylogenetic lineages, elucidating evolutionary mechanisms driving host tropism emergence. Our multidisciplinary work provides a novel molecular basis for how a single, short protein motif could greatly modulate pathogen host tropism.
Organizational Affiliation:
New York State Department of Health, Division of Infectious Diseases, Wadsworth Center, Albany, NY 12208.