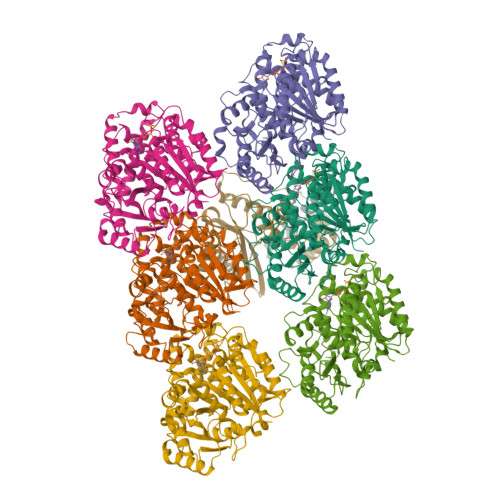
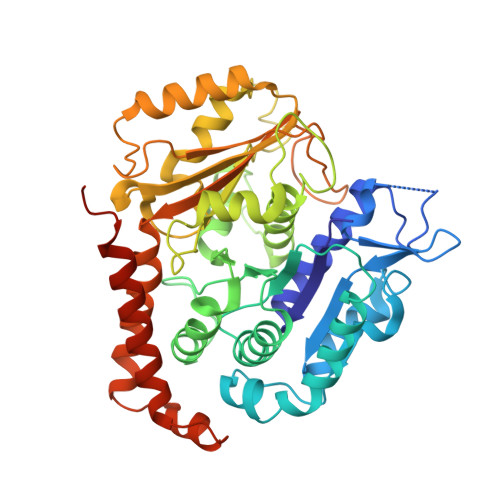
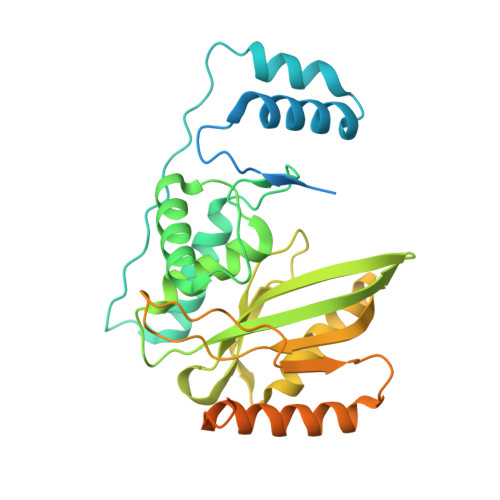
VASH1-SVBP and VASH2-SVBP generate different detyrosination profiles on microtubules.
Ramirez-Rios, S., Choi, S.R., Sanyal, C., Blum, T.B., Bosc, C., Krichen, F., Denarier, E., Soleilhac, J.M., Blot, B., Janke, C., Stoppin-Mellet, V., Magiera, M.M., Arnal, I., Steinmetz, M.O., Moutin, M.J.(2023) J Cell Biol 222
- PubMed: 36512346
- DOI: https://doi.org/10.1083/jcb.202205096
- Primary Citation of Related Structures:
7ZCW - PubMed Abstract:
The detyrosination/tyrosination cycle of α-tubulin is critical for proper cell functioning. VASH1-SVBP and VASH2-SVBP are ubiquitous enzymes involved in microtubule detyrosination, whose mode of action is little known. Here, we show in reconstituted systems and cells that VASH1-SVBP and VASH2-SVBP drive the global and local detyrosination of microtubules, respectively. We solved the cryo-electron microscopy structure of VASH2-SVBP bound to microtubules, revealing a different microtubule-binding configuration of its central catalytic region compared to VASH1-SVBP. We show that the divergent mode of detyrosination between the two enzymes is correlated with the microtubule-binding properties of their disordered N- and C-terminal regions. Specifically, the N-terminal region is responsible for a significantly longer residence time of VASH2-SVBP on microtubules compared to VASH1-SVBP. We suggest that this VASH region is critical for microtubule detachment and diffusion of VASH-SVBP enzymes on lattices. Our results suggest a mechanism by which VASH1-SVBP and VASH2-SVBP could generate distinct microtubule subpopulations and confined areas of detyrosinated lattices to drive various microtubule-based cellular functions.
Organizational Affiliation:
Univ. Grenoble Alpes, Inserm, U1216, Centre National de la Recherche Scientifique, Commissariat à l'Energie Atomique, Grenoble Institut Neurosciences, Grenoble, France.