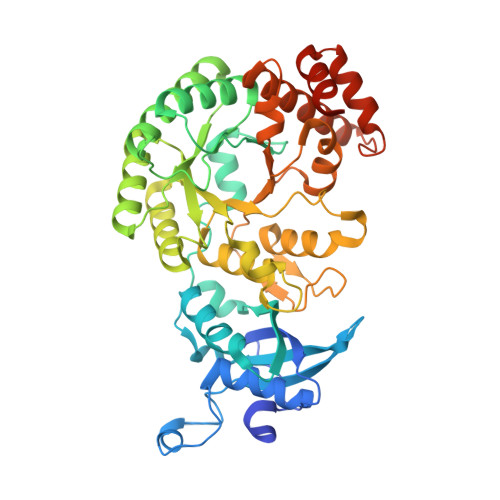
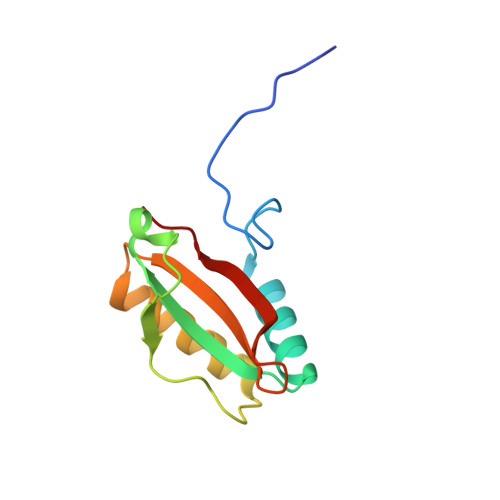
Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Ni, T., Sun, Y., Burn, W., Al-Hazeem, M.M.J., Zhu, Y., Yu, X., Liu, L.N., Zhang, P.(2022) Nat Commun 13: 4299-4299
- PubMed: 35879301
- DOI: https://doi.org/10.1038/s41467-022-32004-w
- Primary Citation of Related Structures:
7ZBT, 7ZC1 - PubMed Abstract:
Carboxysomes are a family of bacterial microcompartments in cyanobacteria and chemoautotrophs. They encapsulate Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) and carbonic anhydrase catalyzing carbon fixation inside a proteinaceous shell. How Rubisco complexes pack within the carboxysomes is unknown. Using cryo-electron tomography, we determine the distinct 3D organization of Rubisco inside two distant α-carboxysomes from a marine α-cyanobacterium Cyanobium sp. PCC 7001 where Rubiscos are organized in three concentric layers, and from a chemoautotrophic bacterium Halothiobacillus neapolitanus where they form intertwining spirals. We further resolve the structures of native Rubisco as well as its higher-order assembly at near-atomic resolutions by subtomogram averaging. The structures surprisingly reveal that the authentic intrinsically disordered linker protein CsoS2 interacts with Rubiscos in native carboxysomes but functions distinctively in the two α-carboxysomes. In contrast to the uniform Rubisco-CsoS2 association in the Cyanobium α-carboxysome, CsoS2 binds only to the Rubiscos close to the shell in the Halo α-carboxysome. Our findings provide critical knowledge of the assembly principles of α-carboxysomes, which may aid in the rational design and repurposing of carboxysome structures for new functions.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.