Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
John, J., Aurelius, O., Srinivas, V., Saura, P., Kim, I.S., Bhowmick, A., Simon, P.S., Dasgupta, M., Pham, C., Gul, S., Sutherlin, K.D., Aller, P., Butryn, A., Orville, A.M., Cheah, M.H., Owada, S., Tono, K., Fuller, F.D., Batyuk, A., Brewster, A.S., Sauter, N.K., Yachandra, V.K., Yano, J., Kaila, V.R.I., Kern, J., Lebrette, H., Hogbom, M.(2022) Elife 11
- PubMed: 36083619
- DOI: https://doi.org/10.7554/eLife.79226
- Primary Citation of Related Structures:
7Z3D, 7Z3E - PubMed Abstract:
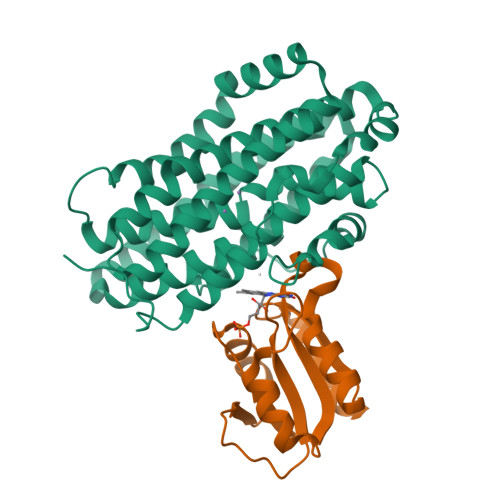
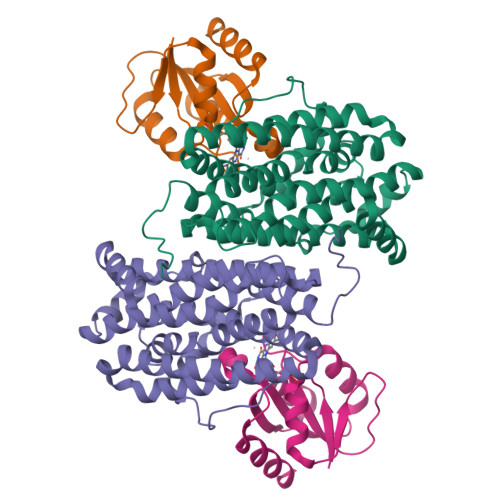
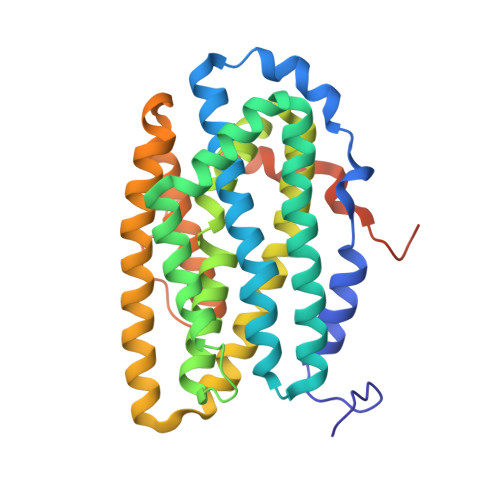
Redox reactions are central to biochemistry and are both controlled by and induce protein structural changes. Here, we describe structural rearrangements and crosstalk within the Bacillus cereus ribonucleotide reductase R2b-NrdI complex, a di-metal carboxylate-flavoprotein system, as part of the mechanism generating the essential catalytic free radical of the enzyme. Femtosecond crystallography at an X-ray free electron laser was utilized to obtain structures at room temperature in defined redox states without suffering photoreduction. Together with density functional theory calculations, we show that the flavin is under steric strain in the R2b-NrdI protein complex, likely tuning its redox properties to promote superoxide generation. Moreover, a binding site in close vicinity to the expected flavin O 2 interaction site is observed to be controlled by the redox state of the flavin and linked to the channel proposed to funnel the produced superoxide species from NrdI to the di-manganese site in protein R2b. These specific features are coupled to further structural changes around the R2b-NrdI interaction surface. The mechanistic implications for the control of reactive oxygen species and radical generation in protein R2b are discussed.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Arrhenius Laboratories for Natural Sciences, Stockholm University, Stockholm, Sweden.