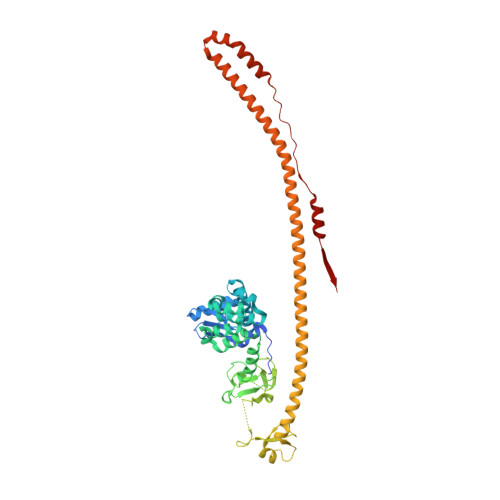
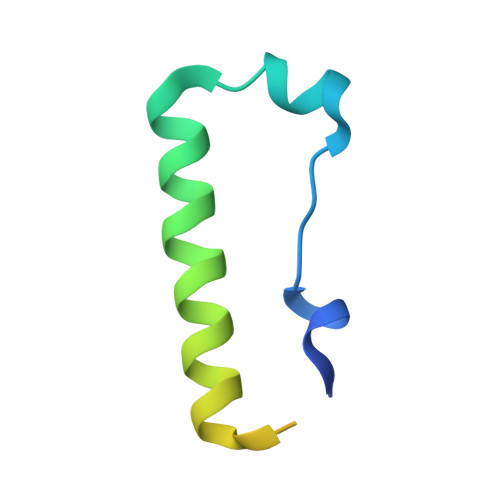
Structure and functional mapping of the KRAB-KAP1 repressor complex.
Stoll, G.A., Pandiloski, N., Douse, C.H., Modis, Y.(2022) EMBO J 41: e111179-e111179
- PubMed: 36341546
- DOI: https://doi.org/10.15252/embj.2022111179
- Primary Citation of Related Structures:
7Z36 - PubMed Abstract:
Transposable elements are a genetic reservoir from which new genes and regulatory elements can emerge. However, expression of transposable elements can be pathogenic and is therefore tightly controlled. KRAB domain-containing zinc finger proteins (KRAB-ZFPs) recruit the co-repressor KRAB-associated protein 1 (KAP1/TRIM28) to regulate many transposable elements, but how KRAB-ZFPs and KAP1 interact remains unclear. Here, we report the crystal structure of the KAP1 tripartite motif (TRIM) in complex with the KRAB domain from a human KRAB-ZFP, ZNF93. Structure-guided mutations in the KAP1-KRAB binding interface abolished repressive activity in an epigenetic transcriptional silencing assay. Deposition of H3K9me3 over thousands of loci is lost genome-wide in cells expressing a KAP1 variant with mutations that abolish KRAB binding. Our work identifies and functionally validates the KRAB-KAP1 molecular interface, which is critical for a central transcriptional control axis in vertebrates. In addition, the structure-based prediction of KAP1 recruitment efficiency will enable optimization of KRABs used in CRISPRi.
Organizational Affiliation:
Molecular Immunity Unit, Department of Medicine, MRC Laboratory of Molecular Biology, University of Cambridge, Cambridge, UK.