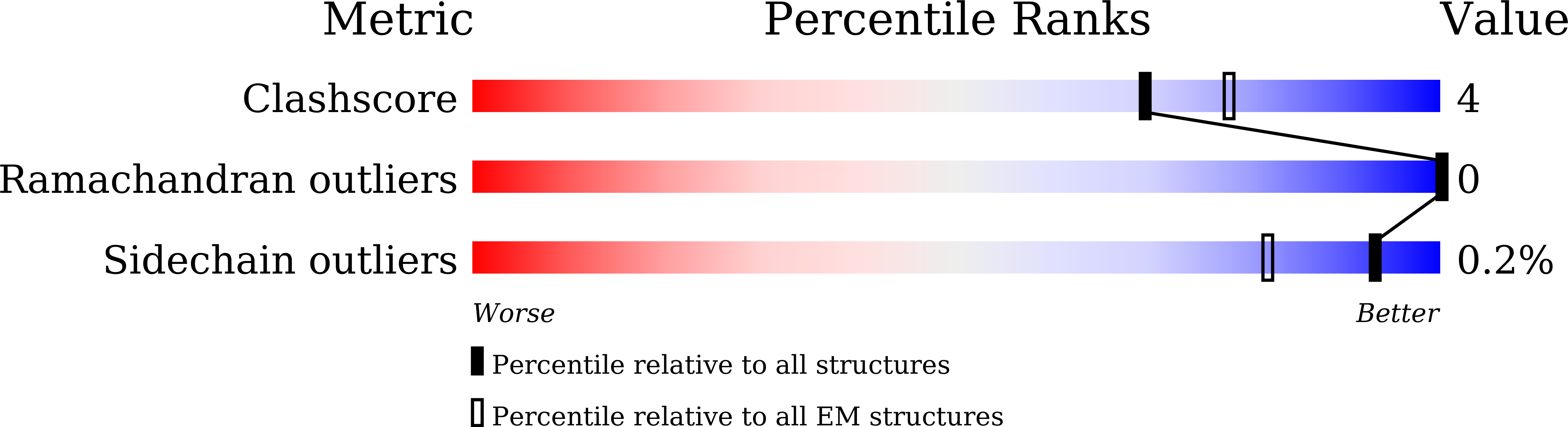Molecular mechanism of antibody neutralization of coxsackievirus A16.
Zhang, C., Liu, C., Shi, J., Wang, Y., Xu, C., Ye, X., Liu, Q., Li, X., Qiao, W., Yin, Y., Cong, Y., Huang, Z.(2022) Nat Commun 13: 7854-7854
- PubMed: 36543790
- DOI: https://doi.org/10.1038/s41467-022-35575-w
- Primary Citation of Related Structures:
7Y7M, 7YMS, 7YRF, 7YRH, 7YV2, 7YV7 - PubMed Abstract:
Coxsackievirus A16 (CVA16) causes hand, foot and mouth disease in infants and young children. However, no vaccine or anti-viral agent is currently available for CVA16. Here, the functions and working mechanisms of two CVA16-specific neutralizing monoclonal antibodies (MAbs), 9B5 and 8C4, are comprehensively investigated. Both 9B5 and 8C4 display potent neutralization in vitro and prophylactic and therapeutic efficacy in a mouse model of CVA16 infection. Mechanistically, 9B5 exerts neutralization primarily through inhibiting CVA16 attachment to cell surface via blockade of CVA16 binding to its attachment receptor, heparan sulfate, whereas 8C4 functions mainly at the post-attachment stage of CVA16 entry by interfering with the interaction between CVA16 and its uncoating receptor SCARB2. Cryo-EM studies show that 9B5 and 8C4 target distinct epitopes located at the 5-fold and 3-fold protrusions of CVA16 capsids, respectively, and exhibit differential binding preference to three forms of naturally occurring CVA16 particles. Moreover, 9B5 and 8C4 are compatible in formulating an antibody cocktail which displays the ability to prevent virus escape seen with individual MAbs. Together, our work elucidates the functional and structural basis of CVA16 antibody-mediated neutralization and protection, providing important information for design and development of effective CVA16 vaccines and antibody therapies.
Organizational Affiliation:
CAS Key Laboratory of Molecular Virology & Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai, China.