Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
Yamanouchi, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calcium-activated potassium channel subunit alpha-1 | 1,060 | Homo sapiens | Mutation(s): 1 Gene Names: KCNMA1 Membrane Entity: Yes | 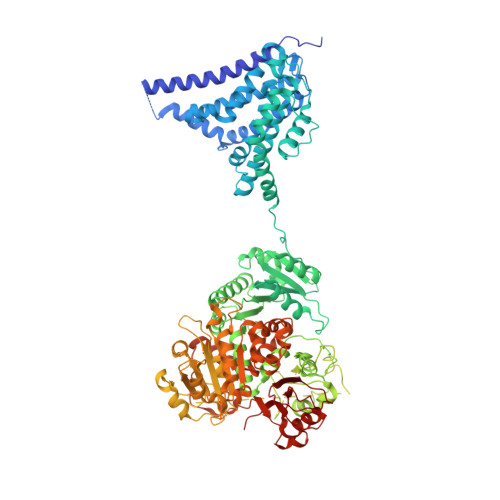 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q12791 (Homo sapiens) Explore Q12791 Go to UniProtKB: Q12791 | |||||
PHAROS: Q12791 GTEx: ENSG00000156113 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12791 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Leucine-rich repeat-containing protein 26 | 334 | Homo sapiens | Mutation(s): 0 Gene Names: LRRC26, CAPC Membrane Entity: Yes | 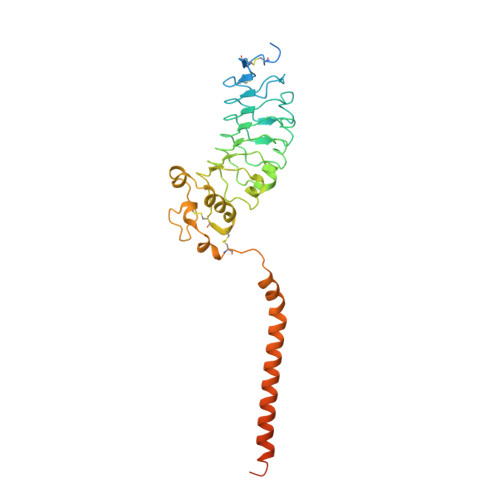 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q2I0M4 (Homo sapiens) Explore Q2I0M4 Go to UniProtKB: Q2I0M4 | |||||
PHAROS: Q2I0M4 GTEx: ENSG00000184709 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2I0M4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CA (Subject of Investigation/LOI) Query on CA | J [auth A] K [auth A] L [auth A] N [auth C] P [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | I [auth A], M [auth C], O [auth E], S [auth G] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | -- |