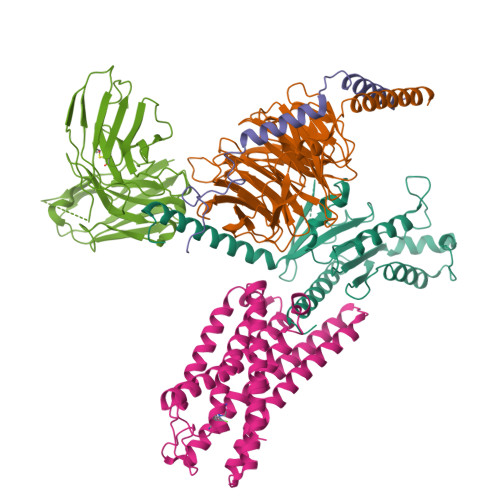
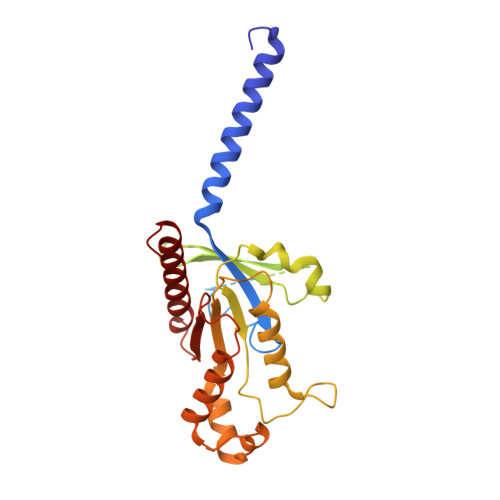
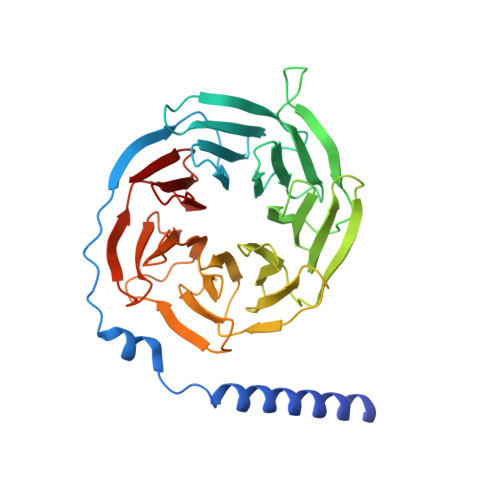
Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Zhao, J., Fu, H., Yu, J., Hong, W., Tian, X., Qi, J., Sun, S., Zhao, C., Wu, C., Xu, Z., Cheng, L., Chai, R., Yan, W., Wei, X., Shao, Z.(2023) Nat Commun 14: 962-962
- PubMed: 36810324
- DOI: https://doi.org/10.1038/s41467-023-36673-z
- Primary Citation of Related Structures:
7YAC, 7YAE - PubMed Abstract:
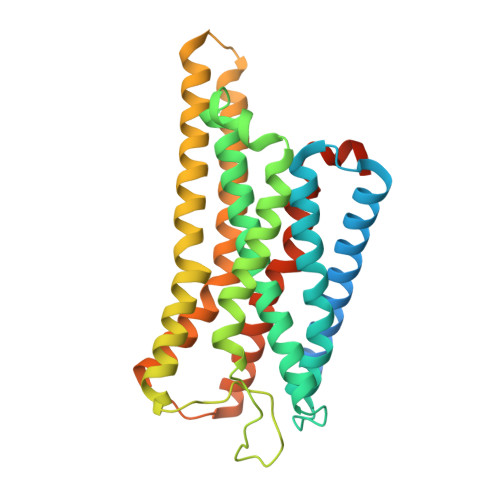
Somatostatin receptor 2 (SSTR2) is highly expressed in neuroendocrine tumors and represents as a therapeutic target. Several peptide analogs mimicking the endogenous ligand somatostatin are available for clinical use, but poor therapeutic effects occur in a subset of patients, which may be correlated with subtype selectivity or cell surface expression. Here, we clarify the signal bias profiles of the first-generation peptide drug octreotide and a new-generation small molecule paltusotine by evaluating their pharmacological characteristics. We then perform cryo-electron microscopy analysis of SSTR2-Gi complexes to determine how the drugs activate SSTR2 in a selective manner. In this work, we decipher the mechanism of ligand recognition, subtype selectivity and signal bias property of SSTR2 sensing octreotide and paltusotine, which may aid in designing therapeutic drugs with specific pharmacological profiles against neuroendocrine tumors.
Organizational Affiliation:
Division of Nephrology and Kidney Research Institute, Laboratory of Aging Research and Cancer Drug Target, State Key Laboratory of Biotherapy and Cancer Center, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan, 610041, China.