Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
Hiragi, K., Nishide, A., Takagi, K., Iwai, K., Kim, M., Mizushima, T.(2023) J Biochem 173: 317-326
- PubMed: 36610722
- DOI: https://doi.org/10.1093/jb/mvac109
- Primary Citation of Related Structures:
7YA7, 7YA8 - PubMed Abstract:
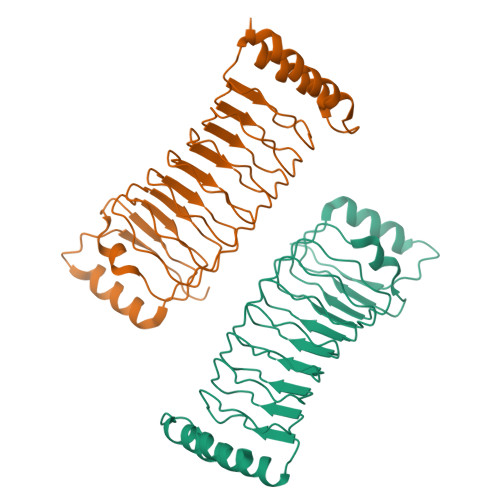
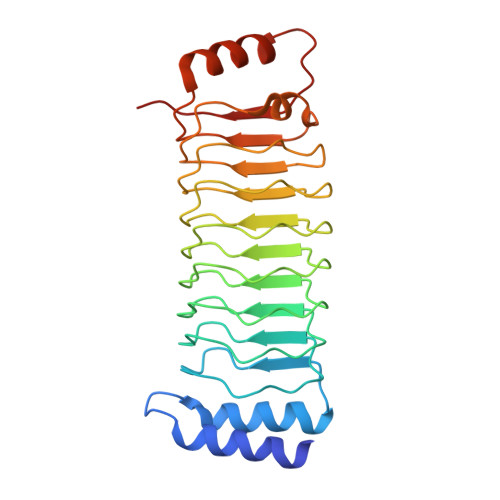
Pathogenic bacteria deliver virulence factors called effectors into host cells in order to facilitate infection. The Shigella effector proteins IpaH1.4 and IpaH2.5 are members of the 'novel E3 ligase' (NEL)-type bacterial E3 ligase family. These proteins ubiquitinate the linear ubiquitin assembly complex (LUBAC) to inhibit nuclear factor (NF)-κB activation and, concomitantly, the inflammatory response. However, the molecular mechanisms underlying the interaction and recognition between IpaH1.4 and IpaH2.5 and LUBAC are unclear. Here we present the crystal structures of the substrate-recognition domains of IpaH1.4 and IpaH2.5 at resolutions of 1.4 and 3.4 Å, respectively. The LUBAC-binding site on IpaH1.4 was predicted based on structural comparisons with the structures of other NEL-type E3s. Structural and biochemical data were collected and analysed to determine the specific residues of IpaH1.4 that are involved in interactions with LUBAC and influence NF-κB signaling. The new structural insight presented here demonstrates how bacterial pathogens target innate immune signaling pathways.
Organizational Affiliation:
Department of Science, Graduate School of Science, University of Hyogo, 2167, Shosha, Himeji, Hyogo, 671-2280, Japan.