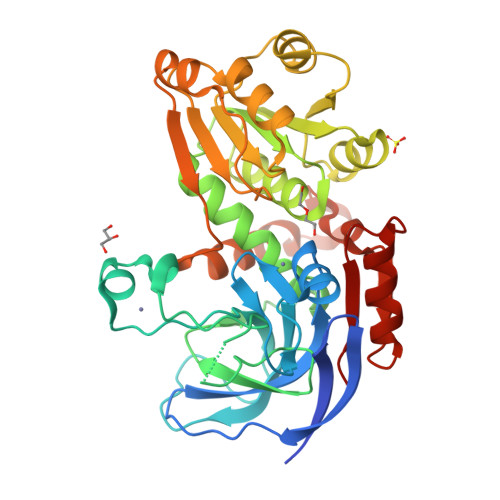
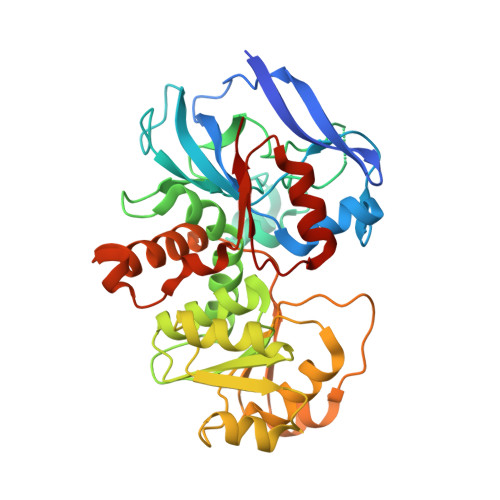
Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Yoshiwara, K., Watanabe, S., Watanabe, Y.(2023) Sci Rep 13: 1920-1920
- PubMed: 36732376
- DOI: https://doi.org/10.1038/s41598-023-29195-7
- Primary Citation of Related Structures:
7Y9P - PubMed Abstract:
Xylitol dehydrogenase (XDH) catalyzes the NAD + -dependent oxidization of xylitol into D-xylulose, and belongs to a zinc-dependent medium-chain dehydrogenase/reductase family. This protein family consists of enzymes with one or two zinc atoms per subunit, among which catalytic zinc is necessary for the activity. Among many XDHs from yeast and fungi, XDH from Pichia stipitis is one of the key enzymes for bioethanol production by lignocellulosic biomass, and possesses only a catalytic zinc atom. Despite its importance in bioindustry, a structural data of XDH has not yet been available, and little insight into the role of a second zinc atom in this protein family is known. We herein report the crystal structure of XDH from P. stipitis using a thermostabilized mutant. In the refined structure, a second zinc atom clearly coordinated with four artificially introduced cysteine ligands. Homologous mutations in XDH from Saccharomyces cerevisiae also stabilized and enhanced activity. The substitution of each of the four cysteine ligands with an aspartate in XDH from Schizosaccharomyces pombe contributed to the significantly better maintenance of activity and thermostability than their substitution with a serine, providing a novel hypothesis for how this zinc atom was eliminated.
Organizational Affiliation:
Department of Bioscience, Graduate School of Agriculture, Ehime University, 3-5-7 Tarumi, Matsuyama, Ehime, 790-8566, Japan.