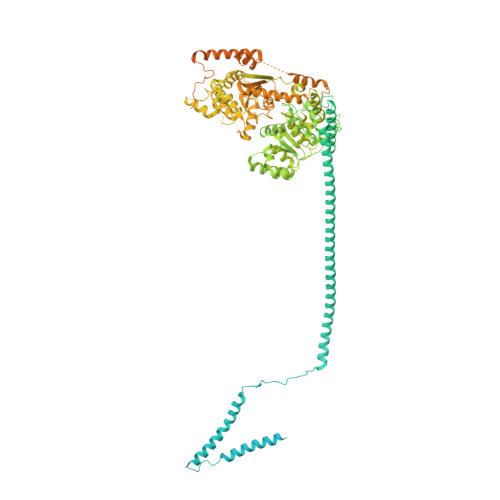
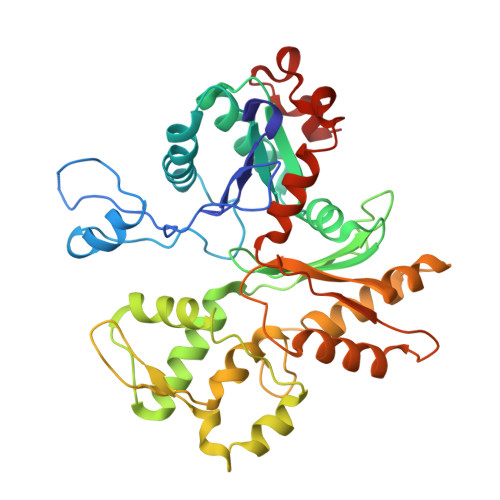
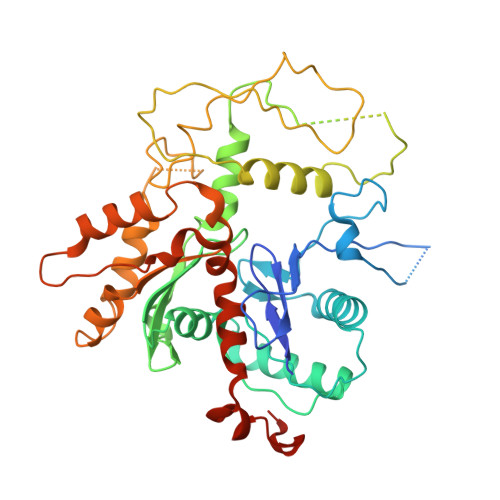
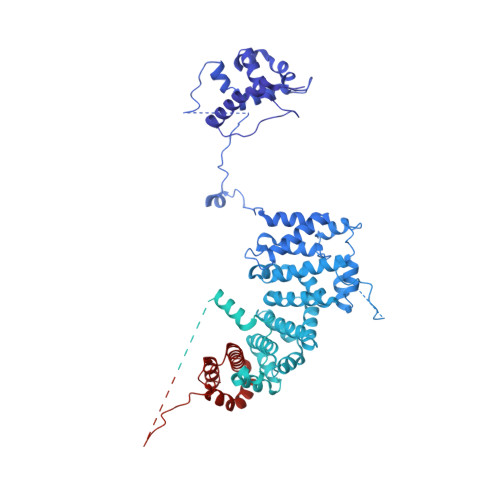
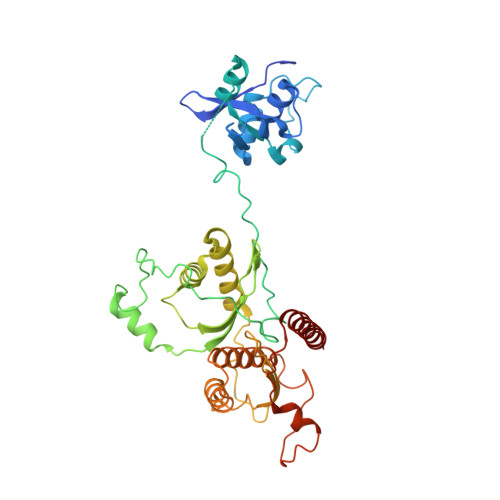
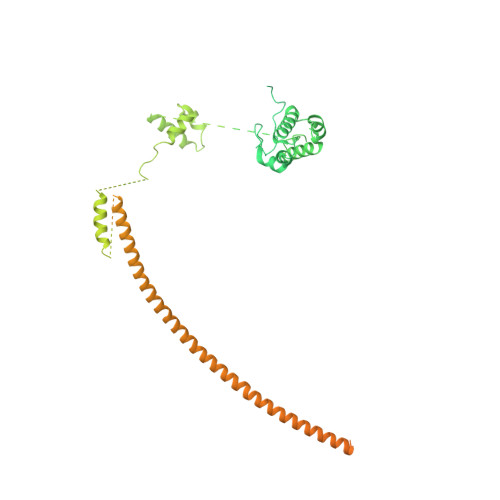
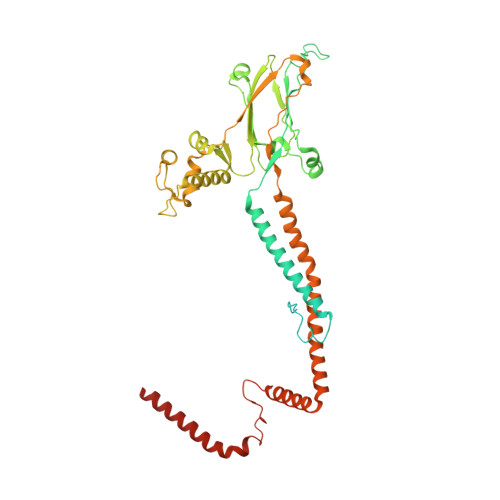
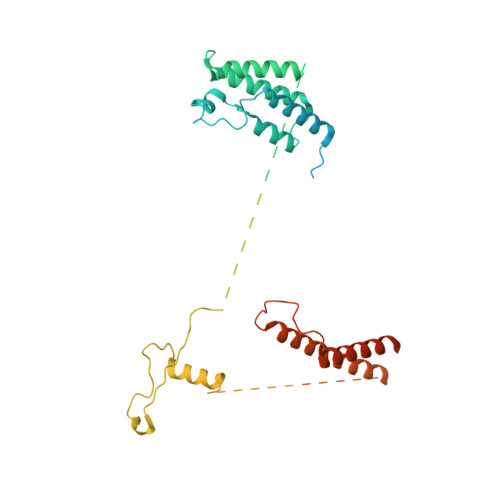
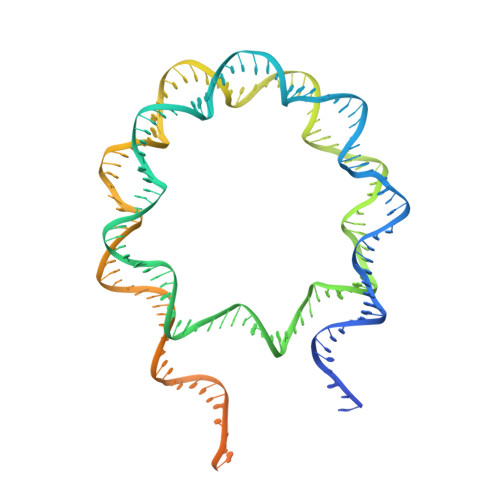
Structure of nucleosome-bound human PBAF complex.
Wang, L., Yu, J., Yu, Z., Wang, Q., Li, W., Ren, Y., Chen, Z., He, S., Xu, Y.(2022) Nat Commun 13: 7644-7644
- PubMed: 36496390
- DOI: https://doi.org/10.1038/s41467-022-34859-5
- Primary Citation of Related Structures:
7Y8R - PubMed Abstract:
BAF and PBAF are mammalian SWI/SNF family chromatin remodeling complexes that possess multiple histone/DNA-binding subunits and create nucleosome-depleted/free regions for transcription activation. Despite previous structural studies and recent advance of SWI/SNF family complexes, it remains incompletely understood how PBAF-nucleosome complex is organized. Here we determined structure of 13-subunit human PBAF in complex with acetylated nucleosome in ADP-BeF 3 -bound state. Four PBAF-specific subunits work together with nine BAF/PBAF-shared subunits to generate PBAF-specific modular organization, distinct from that of BAF at various regions. PBAF-nucleosome structure reveals six histone-binding domains and four DNA-binding domains/modules, the majority of which directly bind histone/DNA. This multivalent nucleosome-binding pattern, not observed in previous studies, suggests that PBAF may integrate comprehensive chromatin information to target genomic loci for function. Our study reveals molecular organization of subunits and histone/DNA-binding domains/modules in PBAF-nucleosome complex and provides structural insights into PBAF-mediated nucleosome association complimentary to the recently reported PBAF-nucleosome structure.
Organizational Affiliation:
Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai, 200032, China.