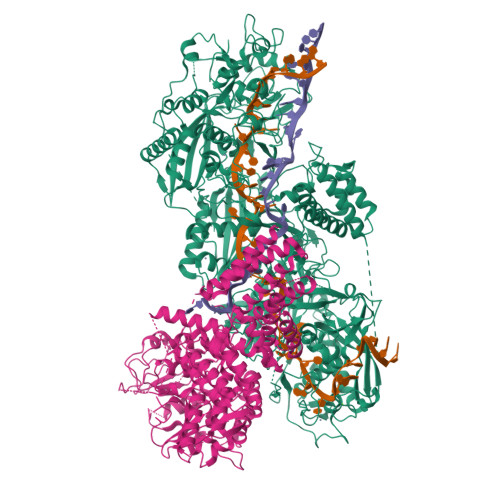
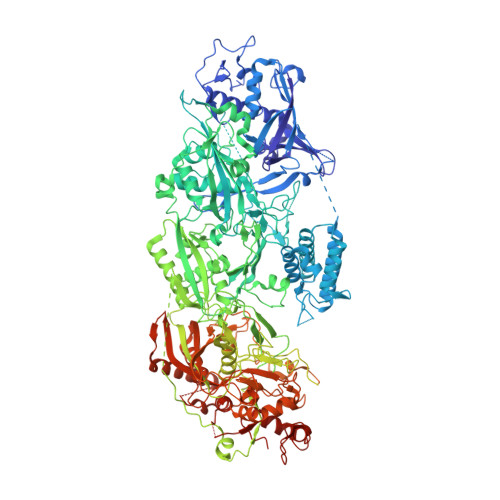
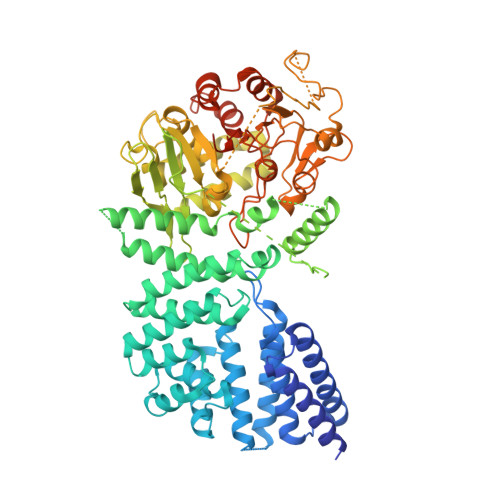
Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Cui, N., Zhang, J.T., Li, Z., Liu, X.Y., Wang, C., Huang, H., Jia, N.(2022) Nat Commun 13: 7549-7549
- PubMed: 36477448
- DOI: https://doi.org/10.1038/s41467-022-35275-5
- Primary Citation of Related Structures:
7Y80, 7Y81, 7Y82, 7Y83, 7Y84, 7Y85 - PubMed Abstract:
The RNA-targeting type III-E CRISPR-gRAMP effector interacts with a caspase-like protease TPR-CHAT to form the CRISPR-guided caspase complex (Craspase), but their functional mechanism is unknown. Here, we report cryo-EM structures of the type III-E gRAMP crRNA and gRAMP crRNA -TPR-CHAT complexes, before and after either self or non-self RNA target binding, and elucidate the mechanisms underlying RNA-targeting and non-self RNA-induced protease activation. The associated TPR-CHAT adopted a distinct conformation upon self versus non-self RNA target binding, with nucleotides at positions -1 and -2 of the CRISPR-derived RNA (crRNA) serving as a sensor. Only binding of the non-self RNA target activated the TPR-CHAT protease, leading to cleavage of Csx30 protein. Furthermore, TPR-CHAT structurally resembled eukaryotic separase, but with a distinct mechanism for protease regulation. Our findings should facilitate the development of gRAMP-based RNA manipulation tools, and advance our understanding of the virus-host discrimination process governed by a nuclease-protease Craspase during type III-E CRISPR-Cas immunity.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Southern University of Science and Technology, Shenzhen, 518055, China.