Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Zhong, Q., Chu, H., Wang, G., Zhang, C., Li, R., Guo, F., Meng, X., Lei, X., Zhou, Y., Ren, R., Tao, L., Li, N., Gao, N., Wei, Y., Qiao, J., Hang, J.(2022) Cell Discov 8: 137-137
- PubMed: 36550107
- DOI: https://doi.org/10.1038/s41421-022-00502-2
- Primary Citation of Related Structures:
7Y5N, 7Y5Q, 8HGG, 8HGH - PubMed Abstract:
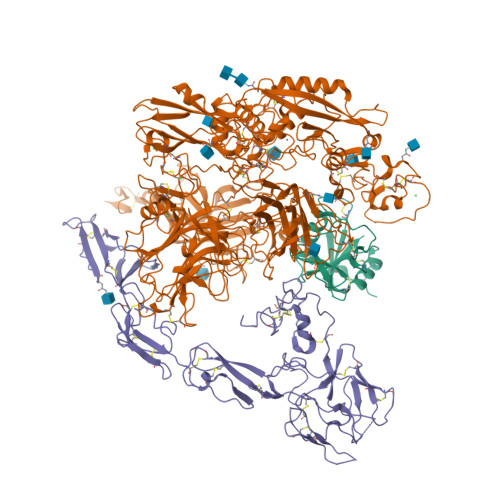
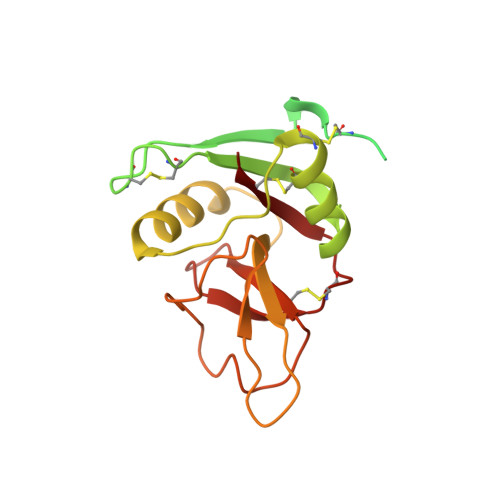
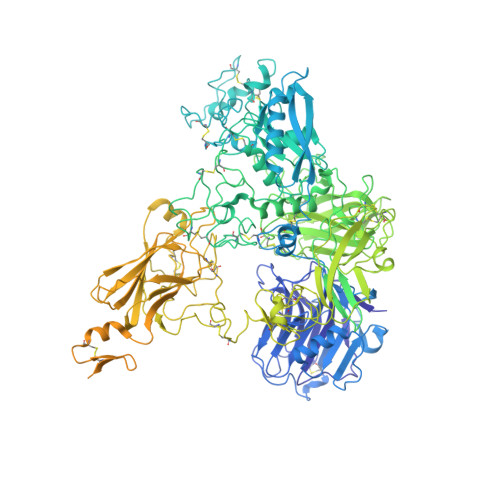
Originally discovered in the circulation of pregnant women as a protein secreted by placental trophoblasts, the metalloprotease pregnancy-associated plasma protein A (PAPP-A) is also widely expressed by many other tissues. It cleaves insulin-like growth factor-binding proteins (IGFBPs) to increase the bioavailability of IGFs and plays essential roles in multiple growth-promoting processes. While the vast majority of the circulatory PAPP-A in pregnancy is proteolytically inactive due to covalent inhibition by proform of eosinophil major basic protein (proMBP), the activity of PAPP-A can also be covalently inhibited by another less characterized modulator, stanniocalcin-2 (STC2). However, the structural basis of PAPP-A proteolysis and the mechanistic differences between these two modulators are poorly understood. Here we present two cryo-EM structures of endogenous purified PAPP-A in complex with either proMBP or STC2. Both modulators form 2:2 heterotetramer with PAPP-A and establish extensive interactions with multiple domains of PAPP-A that are distal to the catalytic cleft. This exosite-binding property results in a steric hindrance to prevent the binding and cleavage of IGFBPs, while the IGFBP linker region-derived peptides harboring the cleavage sites are no longer sensitive to the modulator treatment. Functional investigation into proMBP-mediated PAPP-A regulation in selective intrauterine growth restriction (sIUGR) pregnancy elucidates that PAPP-A and proMBP collaboratively regulate extravillous trophoblast invasion and the consequent fetal growth. Collectively, our work reveals a novel covalent exosite-competitive inhibition mechanism of PAPP-A and its regulatory effect on placental function.
Organizational Affiliation:
Center for Reproductive Medicine, Department of Obstetrics and Gynecology, Peking University Third Hospital, Beijing, China.