Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Sun, N., Huang, J., Qian, J., Zhou, T.P., Guo, J., Tang, L., Zhang, W., Deng, Y., Zhao, W., Wu, G., Liao, R.Z., Chen, X., Zhong, F., Wu, Y.(2022) Nature 611: 715-720
- PubMed: 36130726
- DOI: https://doi.org/10.1038/s41586-022-05342-4
- Primary Citation of Related Structures:
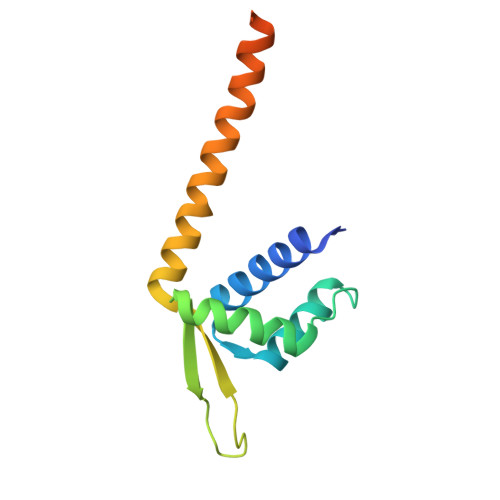
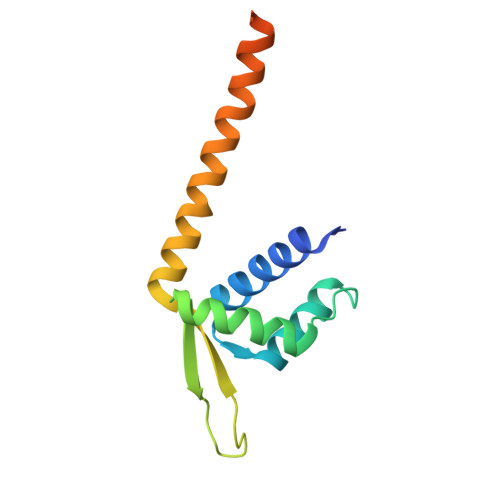
7XUP, 7XUQ - PubMed Abstract:
Naturally evolved enzymes, despite their astonishingly large variety and functional diversity, operate predominantly through thermochemical activation. Integrating prominent photocatalysis modes into proteins, such as triplet energy transfer, could create artificial photoenzymes that expand the scope of natural biocatalysis 1-3 . Here, we exploit genetically reprogrammed, chemically evolved photoenzymes embedded with a synthetic triplet photosensitizer that are capable of excited-state enantio-induction 4-6 . Structural optimization through four rounds of directed evolution afforded proficient variants for the enantioselective intramolecular [2+2]-photocycloaddition of indole derivatives with good substrate generality and excellent enantioselectivities (up to 99% enantiomeric excess). A crystal structure of the photoenzyme-substrate complex elucidated the non-covalent interactions that mediate the reaction stereochemistry. This study expands the energy transfer reactivity 7-10 of artificial triplet photoenzymes in a supramolecular protein cavity and unlocks an integrated approach to valuable enantioselective photochemical synthesis that is not accessible with either the synthetic or the biological world alone.
Organizational Affiliation:
Hubei Engineering Research Center for Biomaterials and Medical Protective Materials, Hubei Key Laboratory of Bioinorganic Chemistry & Materia Medica, Key Laboratory of Material Chemistry for Energy Conversion and Storage, Ministry of Education, School of Chemistry and Chemical Engineering, Huazhong University of Science and Technology (HUST), Wuhan, China.