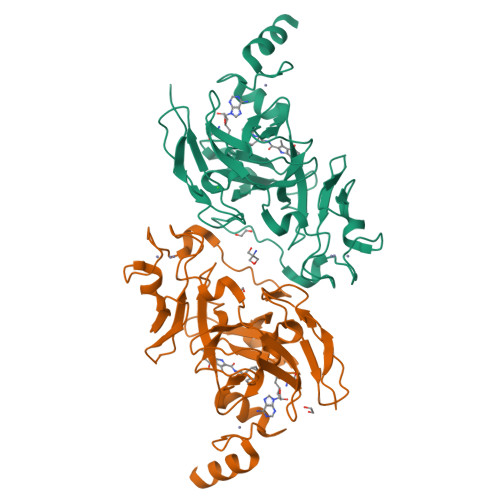
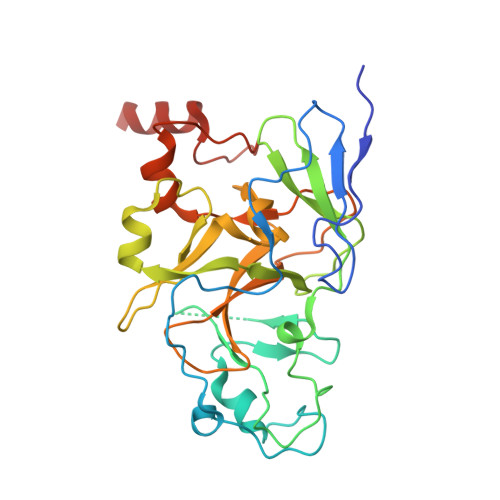
Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
Nishigaya, Y., Takase, S., Sumiya, T., Kikuzato, K., Sato, T., Niwa, H., Sato, S., Nakata, A., Sonoda, T., Hashimoto, N., Namie, R., Honma, T., Umehara, T., Shirouzu, M., Koyama, H., Yoshida, M., Ito, A., Shirai, F.(2023) J Med Chem 66: 4059-4085
- PubMed: 36882960
- DOI: https://doi.org/10.1021/acs.jmedchem.2c02059
- Primary Citation of Related Structures:
7XUA, 7XUB, 7XUC, 7XUD - PubMed Abstract:
Identification of structurally novel inhibitors of lysine methyltransferase G9a has been a subject of intense research in cancer epigenetics. Starting with the high-throughput screening (HTS) hit rac - 10a obtained from the chemical library of the University of Tokyo Drug Discovery Initiative, the structure-activity relationship of the unique substrate-competitive inhibitors was established with the help of X-ray crystallography and fragment molecular orbital (FMO) calculations for the ligand-protein interaction. Further optimization of the in vitro characteristics and drug metabolism and pharmacokinetics (DMPK) properties led to the identification of 26j (RK-701), which is a structurally distinct potent inhibitor of G9a/GLP (IC 50 = 27/53 nM). Compound 26j exhibited remarkable selectivity against other related methyltransferases, dose-dependent attenuation of cellular H3K9me2 levels, and tumor growth inhibition in MOLT-4 cells in vitro . Moreover, compound 26j showed inhibition of tumor initiation and growth in a carcinogen-induced hepatocellular carcinoma (HCC) in vivo mouse model without overt acute toxicity.
Organizational Affiliation:
Watarase Research Center, Discovery Research Headquarters, Kyorin Pharmaceutical Co. Ltd., 1848 Nogi, Shimotsuga-gun, Tochigi 329-0114, Japan.