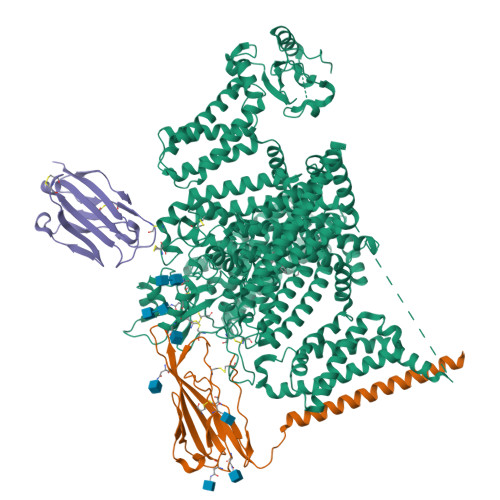
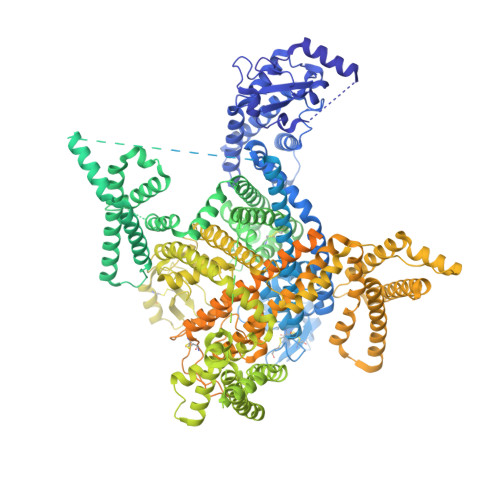
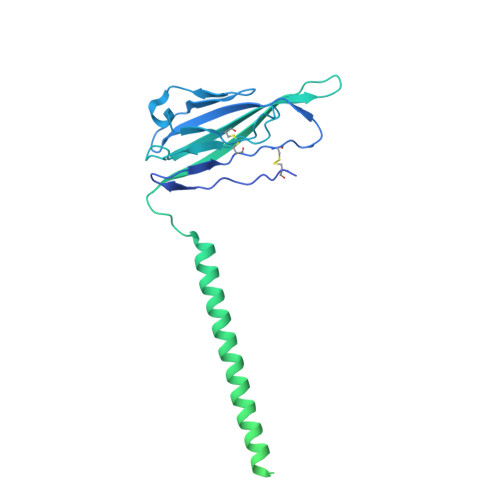
Structural basis for Na V 1.7 inhibition by pore blockers.
Zhang, J., Shi, Y., Huang, Z., Li, Y., Yang, B., Gong, J., Jiang, D.(2022) Nat Struct Mol Biol 29: 1208-1216
- PubMed: 36424527
- DOI: https://doi.org/10.1038/s41594-022-00860-1
- Primary Citation of Related Structures:
7XM9, 7XMF, 7XMG - PubMed Abstract:
Voltage-gated sodium channel Na V 1.7 plays essential roles in pain and odor perception. Na V 1.7 variants cause pain disorders. Accordingly, Na V 1.7 has elicited extensive attention in developing new analgesics. Here we present cryo-EM structures of human Na V 1.7/β1/β2 complexed with inhibitors XEN907, TC-N1752 and Na V 1.7-IN2, explaining specific binding sites and modulation mechanism for the pore blockers. These inhibitors bind in the central cavity blocking ion permeation, but engage different parts of the cavity wall. XEN907 directly causes α- to π-helix transition of DIV-S6 helix, which tightens the fast inactivation gate. TC-N1752 induces π-helix transition of DII-S6 helix mediated by a conserved asparagine on DIII-S6, which closes the activation gate. Na V 1.7-IN2 serves as a pore blocker without causing conformational change. Electrophysiological results demonstrate that XEN907 and TC-N1752 stabilize Na V 1.7 in inactivated state and delay the recovery from inactivation. Our results provide structural framework for Na V 1.7 modulation by pore blockers, and important implications for developing subtype-selective analgesics.
Organizational Affiliation:
Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing, China.