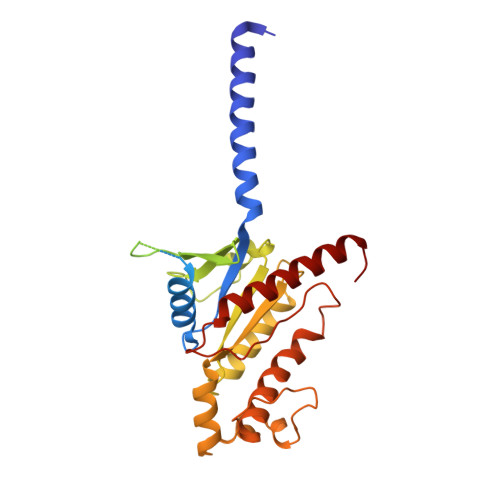
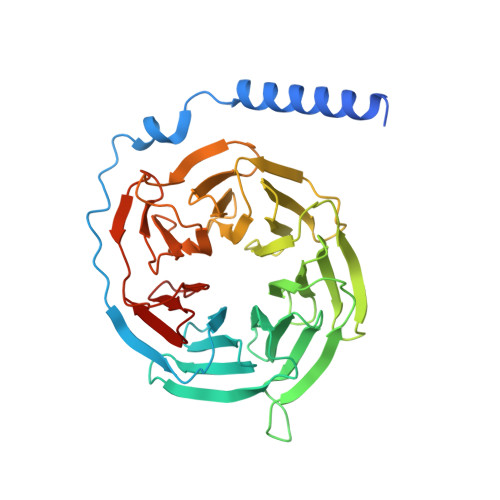
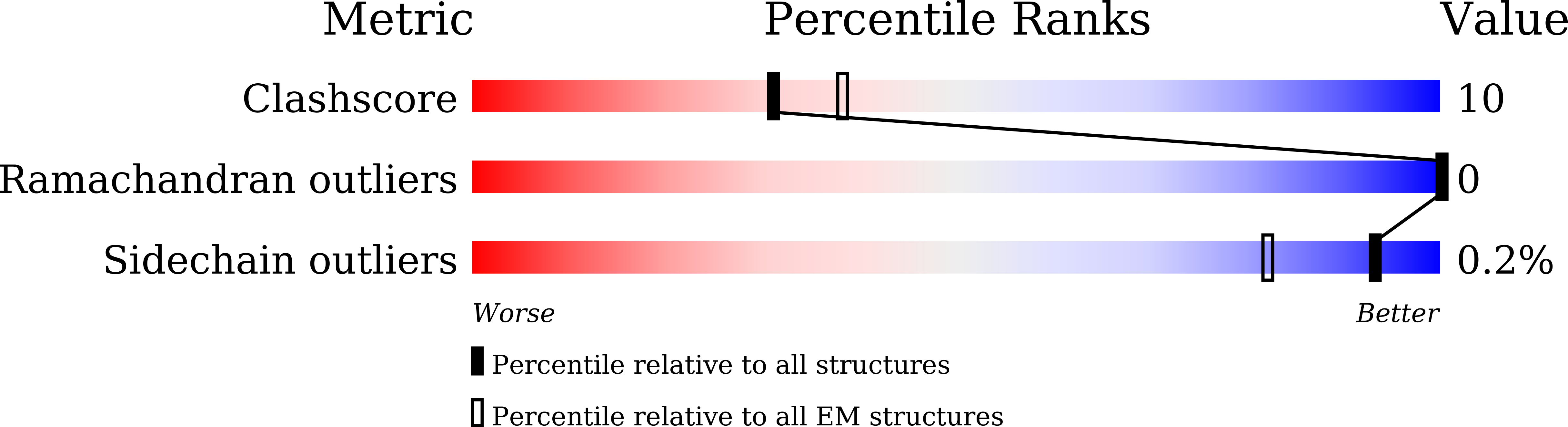Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Lin, H., Xiao, P., Bu, R.Q., Guo, S., Yang, Z., Yuan, D., Zhu, Z.L., Zhang, C.X., He, Q.T., Zhang, C., Ping, Y.Q., Zhao, R.J., Ma, C.S., Liu, C.H., Zhang, X.N., Jiang, D., Huang, S., Xi, Y.T., Zhang, D.L., Xue, C.Y., Yang, B.S., Li, J.Y., Lin, H.C., Zeng, X.H., Zhao, H., Xu, W.M., Yi, F., Liu, Z., Sun, J.P., Yu, X.(2022) Nat Chem Biol 18: 1196-1203
- PubMed: 35982227
- DOI: https://doi.org/10.1038/s41589-022-01084-6
- Primary Citation of Related Structures:
7XKD, 7XKE, 7XKF - PubMed Abstract:
Adhesion G protein-coupled receptors are elusive in terms of their structural information and ligands. Here, we solved the cryogenic-electron microscopy (cryo-EM) structure of apo-ADGRG2, an essential membrane receptor for maintaining male fertility, in complex with a G s trimer. Whereas the formations of two kinks were determinants of the active state, identification of a potential ligand-binding pocket in ADGRG2 facilitated the screening and identification of dehydroepiandrosterone (DHEA), dehydroepiandrosterone sulfate and deoxycorticosterone as potential ligands of ADGRG2. The cryo-EM structures of DHEA-ADGRG2-G s provided interaction details for DHEA within the seven transmembrane domains of ADGRG2. Collectively, our data provide a structural basis for the activation and signaling of ADGRG2, as well as characterization of steroid hormones as ADGRG2 ligands, which might be used as useful tools for further functional studies of the orphan ADGRG2.
Organizational Affiliation:
Key Laboratory of Experimental Teratology of the Ministry of Education, Department of Physiology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, China.