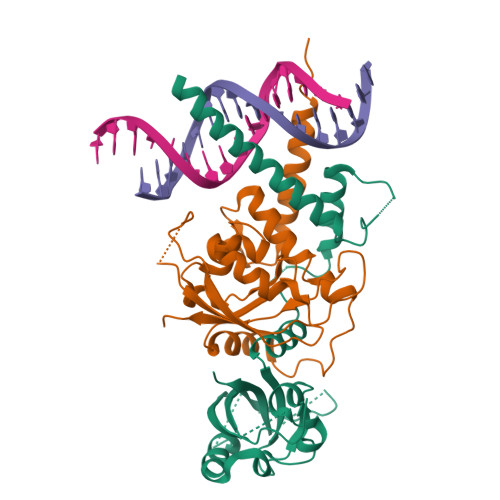
Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Sun, X., Jing, L., Li, F., Zhang, M., Diao, X., Zhuang, J., Rastinejad, F., Wu, D.(2022) Proc Natl Acad Sci U S A 119: e2208804119-e2208804119
- PubMed: 36343253
- DOI: https://doi.org/10.1073/pnas.2208804119
- Primary Citation of Related Structures:
7XHV, 7XI3, 7XI4 - PubMed Abstract:
Neuronal PER-ARNT-SIM (PAS) domain protein 4 (NPAS4) is a protective transcriptional regulator whose dysfunction has been linked to a variety of neuropsychiatric and metabolic diseases. As a member of the basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) transcription factor family, NPAS4 is distinguished by an ability to form functional heterodimers with aryl hydrocarbon receptor nuclear translocator (ARNT) and ARNT2, both of which are also bHLH-PAS family members. Here, we describe the quaternary architectures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers in complexes involving DNA response elements. Our crystallographic studies reveal a uniquely interconnected domain conformation for the NPAS4 protein itself, as well as its differentially configured heterodimeric arrangements with both ARNT and ARNT2. Notably, the PAS-A domains of ARNT and ARNT2 exhibit variable conformations within these two heterodimers. The ARNT PAS-A domain also forms a set of interfaces with the PAS-A and PAS-B domains of NPAS4, different from those previously noted in ARNT heterodimers formed with other class I bHLH-PAS family proteins. Our structural observations together with biochemical and cell-based interrogations of these NPAS4 heterodimers provide molecular glimpses of the NPAS4 protein architecture and extend the known repertoire of heterodimerization patterns within the bHLH-PAS family. The PAS-B domains of NPAS4, ARNT, and ARNT2 all contain ligand-accessible pockets with appropriate volumes required for small-molecule binding. Given NPAS4's linkage to human diseases, the direct visualization of these PAS domains and the further understanding of their relative positioning and interconnections within the NPAS4-ARNT and NPAS4-ARNT2 heterodimers may provide a road map for therapeutic discovery targeting these complexes.
Organizational Affiliation:
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao 266237, China.