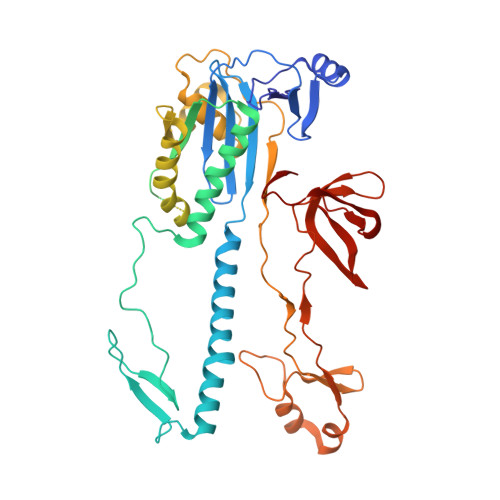
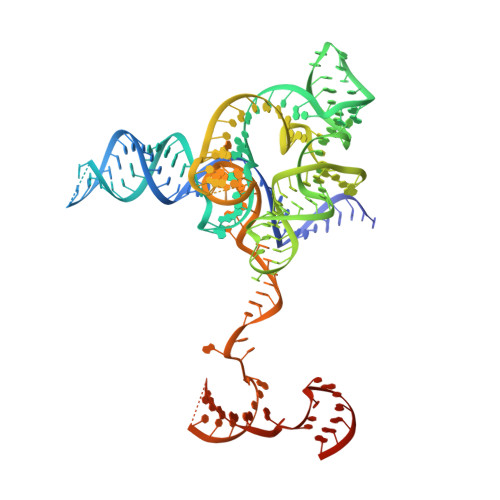
Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Kato, K., Okazaki, S., Kannan, S., Altae-Tran, H., Esra Demircioglu, F., Isayama, Y., Ishikawa, J., Fukuda, M., Macrae, R.K., Nishizawa, T., Makarova, K.S., Koonin, E.V., Zhang, F., Nishimasu, H.(2022) Nat Commun 13: 6719-6719
- PubMed: 36344504
- DOI: https://doi.org/10.1038/s41467-022-34378-3
- Primary Citation of Related Structures:
7XHT - PubMed Abstract:
Transposon-encoded IscB family proteins are RNA-guided nucleases in the OMEGA (obligate mobile element-guided activity) system, and likely ancestors of the RNA-guided nuclease Cas9 in the type II CRISPR-Cas adaptive immune system. IscB associates with its cognate ωRNA to form a ribonucleoprotein complex that cleaves double-stranded DNA targets complementary to an ωRNA guide segment. Although IscB shares the RuvC and HNH endonuclease domains with Cas9, it is much smaller than Cas9, mainly due to the lack of the α-helical nucleic-acid recognition lobe. Here, we report the cryo-electron microscopy structure of an IscB protein from the human gut metagenome (OgeuIscB) in complex with its cognate ωRNA and a target DNA, at 2.6-Å resolution. This high-resolution structure reveals the detailed architecture of the IscB-ωRNA ribonucleoprotein complex, and shows how the small IscB protein assembles with the ωRNA and mediates RNA-guided DNA cleavage. The large ωRNA scaffold structurally and functionally compensates for the recognition lobe of Cas9, and participates in the recognition of the guide RNA-target DNA heteroduplex. These findings provide insights into the mechanism of the programmable DNA cleavage by the IscB-ωRNA complex and the evolution of the type II CRISPR-Cas9 effector complexes.
Organizational Affiliation:
Structural Biology Division, Research Center for Advanced Science and Technology, The University of Tokyo, Tokyo, Japan.