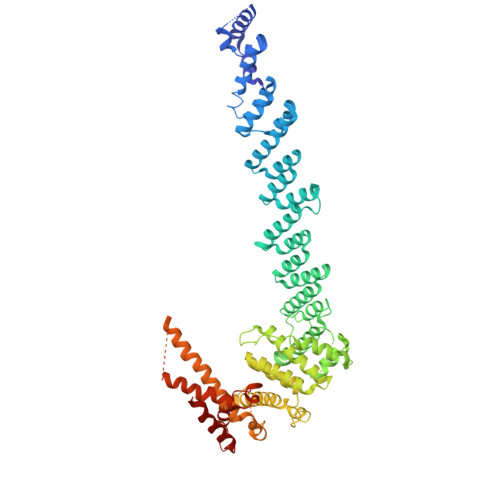
Structural basis for Gemin5 decamer-mediated mRNA binding.
Guo, Q., Zhao, S., Francisco-Velilla, R., Zhang, J., Embarc-Buh, A., Abellan, S., Lv, M., Tang, P., Gong, Q., Shen, H., Sun, L., Yao, X., Min, J., Shi, Y., Martinez-Salas, E., Zhang, K., Xu, C.(2022) Nat Commun 13: 5166-5166
- PubMed: 36056043
- DOI: https://doi.org/10.1038/s41467-022-32883-z
- Primary Citation of Related Structures:
7XDT, 7XGR - PubMed Abstract:
Gemin5 in the Survival Motor Neuron (SMN) complex serves as the RNA-binding protein to deliver small nuclear RNAs (snRNAs) to the small nuclear ribonucleoprotein Sm complex via its N-terminal WD40 domain. Additionally, the C-terminal region plays an important role in regulating RNA translation by directly binding to viral RNAs and cellular mRNAs. Here, we present the three-dimensional structure of the Gemin5 C-terminal region, which adopts a homodecamer architecture comprised of a dimer of pentamers. By structural analysis, mutagenesis, and RNA-binding assays, we find that the intact pentamer/decamer is critical for the Gemin5 C-terminal region to bind cognate RNA ligands and to regulate mRNA translation. The Gemin5 high-order architecture is assembled via pentamerization, allowing binding to RNA ligands in a coordinated manner. We propose a model depicting the regulatory role of Gemin5 in selective RNA binding and translation. Therefore, our work provides insights into the SMN complex-independent function of Gemin5.
Organizational Affiliation:
MOE Key Laboratory for Cellular Dynamics, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, 230027, Hefei, China.