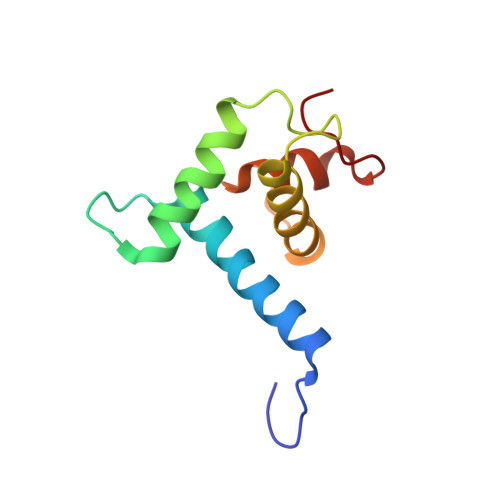
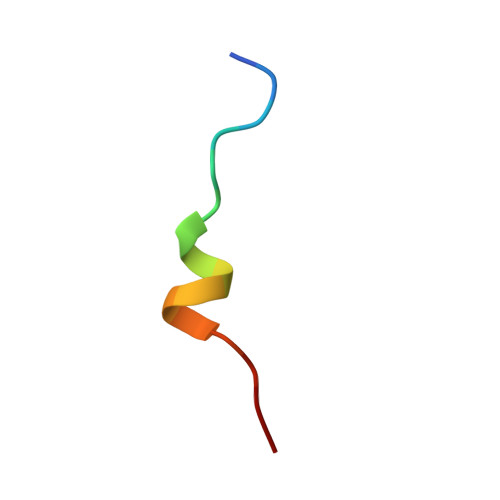
Structural mechanism of BRD4-NUT and p300 bipartite interaction in propagating aberrant gene transcription in chromatin in NUT carcinoma.
Yu, D., Liang, Y., Kim, C., Jaganathan, A., Ji, D., Han, X., Yang, X., Jia, Y., Gu, R., Wang, C., Zhang, Q., Cheung, K.L., Zhou, M.M., Zeng, L.(2023) Nat Commun 14: 378-378
- PubMed: 36690674
- DOI: https://doi.org/10.1038/s41467-023-36063-5
- Primary Citation of Related Structures:
7XEZ, 7XFG - PubMed Abstract:
BRD4-NUT, a driver fusion mutant in rare and highly aggressive NUT carcinoma, acts in aberrant transcription of anti-differentiation genes by recruiting histone acetyltransferase (HAT) p300 and promoting p300-driven histone hyperacetylation and nuclear condensation in chromatin. However, the molecular basis of how BRD4-NUT recruits and activates p300 remains elusive. Here, we report that BRD4-NUT contains two transactivation domains (TADs) in NUT that bind to the TAZ2 domain in p300. Our NMR structures reveal that NUT TADs adopt amphipathic helices when bound to the four-helical bundle TAZ2 domain. The NUT protein forms liquid-like droplets in-vitro that are enhanced by TAZ2 binding in 1:2 stoichiometry. The TAD/TAZ2 bipartite binding in BRD4-NUT/p300 triggers allosteric activation of p300 and acetylation-driven liquid-like condensation on chromatin that comprise histone H3 lysine 27 and 18 acetylation and transcription proteins BRD4L/S, CDK9, MED1, and RNA polymerase II. The BRD4-NUT/p300 chromatin condensation is key for activating transcription of pro-proliferation genes such as ALX1, resulting ALX1/Snail signaling and epithelial-to-mesenchymal transition. Our study provides a previously underappreciated structural mechanism illuminating BRD4-NUT's bipartite p300 recruitment and activation in NUT carcinoma that nucleates a feed-forward loop for propagating histone hyperacetylation and chromatin condensation to sustain aberrant anti-differentiation gene transcription and perpetual tumor cell growth.
Organizational Affiliation:
Bethune Institute of Epigenetic Medicine, The First Hospital of Jilin University, Changchun, Jilin, 130021, China.