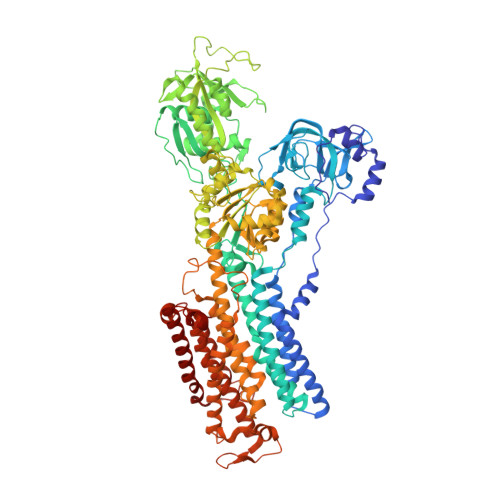
Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Young, V.C., Nakanishi, H., Meyer, D.J., Nishizawa, T., Oshima, A., Artigas, P., Abe, K.(2022) Nat Commun 13: 5270-5270
- PubMed: 36085139
- DOI: https://doi.org/10.1038/s41467-022-32793-0
- Primary Citation of Related Structures:
7X20, 7X21, 7X22, 7X23, 7X24 - PubMed Abstract:
Ion-transport mechanisms evolve by changing ion-selectivity, such as switching from Na + to H + selectivity in secondary-active transporters or P-type-ATPases. Here we study primary-active transport via P-type ATPases using functional and structural analyses to demonstrate that four simultaneous residue substitutions transform the non-gastric H + /K + pump, a strict H + -dependent electroneutral P-type ATPase, into a bona fide Na + -dependent electrogenic Na + /K + pump. Conversion of a H + -dependent primary-active transporter into a Na + -dependent one provides a prototype for similar studies of ion-transport proteins. Moreover, we solve the structures of the wild-type non-gastric H + /K + pump, a suitable drug target to treat cystic fibrosis, and of its Na + /K + pump-mimicking mutant in two major conformations, providing insight on how Na + binding drives a concerted mechanism leading to Na + /K + pump phosphorylation.
Organizational Affiliation:
Department of Cell Physiology and Molecular Biophysics, Center for Membrane Protein Research, Texas Tech University Health Sciences Center, Lubbock, TX, USA.